Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
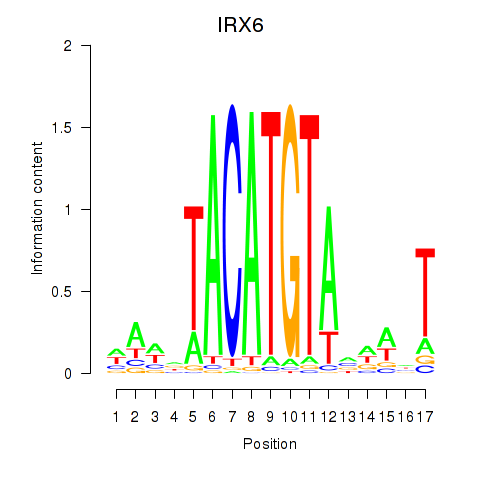
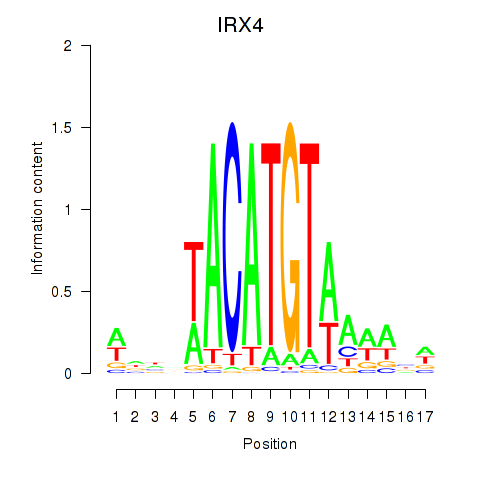
Results for IRX6_IRX4
Z-value: 0.39
Transcription factors associated with IRX6_IRX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX6
|
ENSG00000159387.7 | iroquois homeobox 6 |
|
IRX4
|
ENSG00000113430.5 | iroquois homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX4 | hg19_v2_chr5_-_1882858_1883003 | -0.33 | 6.7e-01 | Click! |
| IRX6 | hg19_v2_chr16_+_55357672_55357672 | 0.19 | 8.1e-01 | Click! |
Activity profile of IRX6_IRX4 motif
Sorted Z-values of IRX6_IRX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_201438282 | 0.44 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr16_+_776936 | 0.25 |
ENST00000549114.1
ENST00000341413.4 ENST00000562187.1 ENST00000564537.1 |
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr19_-_54974894 | 0.20 |
ENST00000333834.4
|
LENG9
|
leukocyte receptor cluster (LRC) member 9 |
| chr7_-_55583740 | 0.16 |
ENST00000453256.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr16_+_777739 | 0.16 |
ENST00000563792.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr16_+_777246 | 0.13 |
ENST00000561546.1
ENST00000564545.1 ENST00000389703.3 ENST00000567414.1 ENST00000568141.1 |
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr3_-_9994021 | 0.13 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr1_-_3566627 | 0.10 |
ENST00000419924.2
ENST00000270708.7 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr3_+_186158169 | 0.10 |
ENST00000435548.1
ENST00000421006.1 |
RP11-78H24.1
|
RP11-78H24.1 |
| chr16_+_777118 | 0.09 |
ENST00000562141.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr20_+_43990576 | 0.09 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr1_-_3566590 | 0.08 |
ENST00000424367.1
ENST00000378322.3 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr3_-_124606502 | 0.08 |
ENST00000483168.1
|
ITGB5
|
integrin, beta 5 |
| chr13_-_46716969 | 0.08 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr7_+_871559 | 0.07 |
ENST00000421580.1
|
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr15_+_41057818 | 0.07 |
ENST00000558467.1
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr2_-_231989808 | 0.07 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr2_+_58655461 | 0.06 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr11_+_114166536 | 0.06 |
ENST00000299964.3
|
NNMT
|
nicotinamide N-methyltransferase |
| chr6_-_33679452 | 0.06 |
ENST00000374231.4
ENST00000607484.1 ENST00000374214.3 |
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr11_+_110225855 | 0.05 |
ENST00000526605.1
ENST00000526703.1 |
RP11-347E10.1
|
RP11-347E10.1 |
| chr18_-_56985776 | 0.05 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr3_+_53528659 | 0.05 |
ENST00000350061.5
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr1_-_149859466 | 0.05 |
ENST00000331128.3
|
HIST2H2AB
|
histone cluster 2, H2ab |
| chr18_-_46784778 | 0.05 |
ENST00000582399.1
|
DYM
|
dymeclin |
| chr18_+_616672 | 0.05 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr8_+_84824920 | 0.04 |
ENST00000523678.1
|
RP11-120I21.2
|
RP11-120I21.2 |
| chr17_-_42327236 | 0.04 |
ENST00000399246.2
|
AC003102.1
|
AC003102.1 |
| chr1_-_89641680 | 0.04 |
ENST00000294671.2
|
GBP7
|
guanylate binding protein 7 |
| chr7_+_23210760 | 0.04 |
ENST00000366347.4
|
AC005082.1
|
Uncharacterized protein |
| chr4_+_144312659 | 0.04 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chrX_-_15333736 | 0.04 |
ENST00000380470.3
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr4_-_153303658 | 0.03 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr2_-_183106641 | 0.03 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chrX_+_15767971 | 0.03 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr8_+_12803176 | 0.03 |
ENST00000524591.2
|
KIAA1456
|
KIAA1456 |
| chr5_-_177207634 | 0.03 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chr1_-_152086556 | 0.03 |
ENST00000368804.1
|
TCHH
|
trichohyalin |
| chr12_+_53848549 | 0.02 |
ENST00000439930.3
ENST00000548933.1 ENST00000562264.1 |
PCBP2
|
poly(rC) binding protein 2 |
| chr5_+_59783540 | 0.02 |
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr6_-_117747015 | 0.02 |
ENST00000368508.3
ENST00000368507.3 |
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase |
| chr11_+_61891445 | 0.02 |
ENST00000394818.3
ENST00000533896.1 ENST00000278849.4 |
INCENP
|
inner centromere protein antigens 135/155kDa |
| chrX_+_99899180 | 0.02 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr1_+_67673297 | 0.02 |
ENST00000425614.1
ENST00000395227.1 |
IL23R
|
interleukin 23 receptor |
| chr7_-_16844611 | 0.02 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr10_+_118349920 | 0.02 |
ENST00000531984.1
|
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr7_-_92855762 | 0.02 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr17_-_47723943 | 0.01 |
ENST00000510476.1
ENST00000503676.1 |
SPOP
|
speckle-type POZ protein |
| chr12_-_10282836 | 0.01 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr5_+_175490540 | 0.01 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr3_+_111758553 | 0.01 |
ENST00000419127.1
|
TMPRSS7
|
transmembrane protease, serine 7 |
| chr12_+_32655110 | 0.01 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr14_-_68159152 | 0.01 |
ENST00000554035.1
|
RDH11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr1_+_170904612 | 0.01 |
ENST00000367759.4
ENST00000367758.3 |
MROH9
|
maestro heat-like repeat family member 9 |
| chr3_-_168865522 | 0.01 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr16_+_21689835 | 0.01 |
ENST00000286149.4
ENST00000388958.3 |
OTOA
|
otoancorin |
| chr1_-_110284384 | 0.01 |
ENST00000540225.1
|
GSTM3
|
glutathione S-transferase mu 3 (brain) |
| chr12_-_10282742 | 0.01 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr4_+_123653882 | 0.01 |
ENST00000433287.1
|
BBS12
|
Bardet-Biedl syndrome 12 |
| chr17_-_6524159 | 0.01 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr4_+_40194609 | 0.01 |
ENST00000508513.1
|
RHOH
|
ras homolog family member H |
| chr4_+_40194570 | 0.00 |
ENST00000507851.1
|
RHOH
|
ras homolog family member H |
| chr17_-_29641084 | 0.00 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chrX_-_135962923 | 0.00 |
ENST00000565438.1
|
RBMX
|
RNA binding motif protein, X-linked |
| chr1_-_8763278 | 0.00 |
ENST00000468247.1
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr8_-_101724989 | 0.00 |
ENST00000517403.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr19_+_50691437 | 0.00 |
ENST00000598205.1
|
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr21_-_48024986 | 0.00 |
ENST00000291700.4
ENST00000367071.4 |
S100B
|
S100 calcium binding protein B |
| chr8_-_74495065 | 0.00 |
ENST00000523533.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr12_-_124118151 | 0.00 |
ENST00000534960.1
|
EIF2B1
|
eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa |
| chr1_+_212475148 | 0.00 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX6_IRX4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |