Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
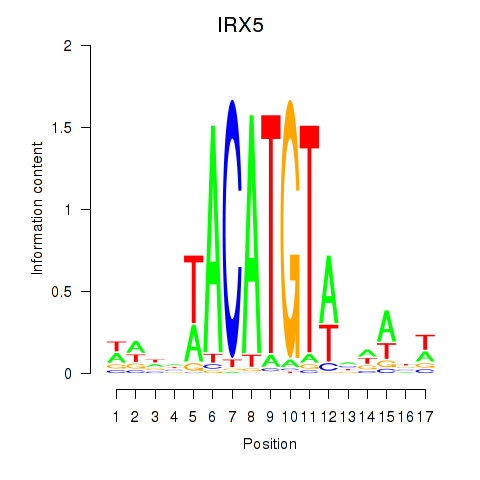
Results for IRX5
Z-value: 0.40
Transcription factors associated with IRX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX5
|
ENSG00000176842.10 | iroquois homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX5 | hg19_v2_chr16_+_54964740_54964789 | 0.74 | 2.6e-01 | Click! |
Activity profile of IRX5 motif
Sorted Z-values of IRX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_149859466 | 0.22 |
ENST00000331128.3
|
HIST2H2AB
|
histone cluster 2, H2ab |
| chr7_+_23210760 | 0.13 |
ENST00000366347.4
|
AC005082.1
|
Uncharacterized protein |
| chr8_+_84824920 | 0.12 |
ENST00000523678.1
|
RP11-120I21.2
|
RP11-120I21.2 |
| chr3_+_186158169 | 0.10 |
ENST00000435548.1
ENST00000421006.1 |
RP11-78H24.1
|
RP11-78H24.1 |
| chr4_+_189376725 | 0.10 |
ENST00000510832.1
ENST00000510005.1 ENST00000503458.1 |
LINC01060
|
long intergenic non-protein coding RNA 1060 |
| chr2_+_58655461 | 0.09 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr12_+_56114151 | 0.09 |
ENST00000547072.1
ENST00000552930.1 ENST00000257895.5 |
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr13_-_46716969 | 0.09 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr9_+_95909309 | 0.08 |
ENST00000366188.2
|
RP11-370F5.4
|
RP11-370F5.4 |
| chr11_-_1619524 | 0.08 |
ENST00000412090.1
|
KRTAP5-2
|
keratin associated protein 5-2 |
| chr6_+_12008986 | 0.08 |
ENST00000491710.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr8_+_101349823 | 0.07 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr15_+_41057818 | 0.06 |
ENST00000558467.1
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr1_+_81106951 | 0.06 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr5_+_140566 | 0.06 |
ENST00000502646.1
|
PLEKHG4B
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4B |
| chr13_-_34250861 | 0.06 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr4_+_144312659 | 0.06 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr18_-_69246167 | 0.06 |
ENST00000566582.1
|
RP11-510D19.1
|
RP11-510D19.1 |
| chr12_+_56114189 | 0.05 |
ENST00000548082.1
|
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chrX_-_52258669 | 0.05 |
ENST00000441417.1
|
XAGE1A
|
X antigen family, member 1A |
| chrX_+_52513455 | 0.05 |
ENST00000446098.1
|
XAGE1C
|
X antigen family, member 1C |
| chr6_+_26204825 | 0.05 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr12_+_32655110 | 0.05 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr1_-_152086556 | 0.05 |
ENST00000368804.1
|
TCHH
|
trichohyalin |
| chr12_-_11036844 | 0.05 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr8_+_9009202 | 0.04 |
ENST00000518496.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr3_+_52245458 | 0.04 |
ENST00000459884.1
|
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chrX_+_153238220 | 0.04 |
ENST00000425274.1
|
TMEM187
|
transmembrane protein 187 |
| chr4_+_123653882 | 0.04 |
ENST00000433287.1
|
BBS12
|
Bardet-Biedl syndrome 12 |
| chr5_-_13944652 | 0.04 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chrX_+_99899180 | 0.04 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr12_+_21679220 | 0.04 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr4_-_22444733 | 0.04 |
ENST00000508133.1
|
GPR125
|
G protein-coupled receptor 125 |
| chr4_+_141264597 | 0.04 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr12_+_2912363 | 0.04 |
ENST00000544366.1
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr17_+_12859080 | 0.04 |
ENST00000583608.1
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr20_+_10199468 | 0.04 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr2_-_127963343 | 0.04 |
ENST00000335247.7
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr11_+_110225855 | 0.04 |
ENST00000526605.1
ENST00000526703.1 |
RP11-347E10.1
|
RP11-347E10.1 |
| chr13_+_24844857 | 0.03 |
ENST00000409126.1
ENST00000343003.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr19_-_36909528 | 0.03 |
ENST00000392161.3
ENST00000392171.1 |
ZFP82
|
ZFP82 zinc finger protein |
| chr11_-_31014214 | 0.03 |
ENST00000406071.2
ENST00000339794.5 |
DCDC1
|
doublecortin domain containing 1 |
| chrX_+_52240504 | 0.03 |
ENST00000399805.2
|
XAGE1B
|
X antigen family, member 1B |
| chr19_-_52227221 | 0.03 |
ENST00000222115.1
ENST00000540069.2 |
HAS1
|
hyaluronan synthase 1 |
| chr22_+_20877924 | 0.03 |
ENST00000445189.1
|
MED15
|
mediator complex subunit 15 |
| chr2_+_173686303 | 0.03 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr18_-_69246186 | 0.03 |
ENST00000568095.1
|
RP11-510D19.1
|
RP11-510D19.1 |
| chr11_-_60010556 | 0.03 |
ENST00000427611.2
|
MS4A4E
|
membrane-spanning 4-domains, subfamily A, member 4E |
| chr2_+_38177620 | 0.03 |
ENST00000402091.3
|
RMDN2
|
regulator of microtubule dynamics 2 |
| chr10_-_14050522 | 0.03 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr1_+_10509971 | 0.03 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr12_-_10282836 | 0.03 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr7_-_111424506 | 0.03 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr11_+_61891445 | 0.03 |
ENST00000394818.3
ENST00000533896.1 ENST00000278849.4 |
INCENP
|
inner centromere protein antigens 135/155kDa |
| chr2_+_38177575 | 0.03 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr4_+_169842707 | 0.03 |
ENST00000503290.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr18_+_33709834 | 0.03 |
ENST00000358232.6
ENST00000351393.6 ENST00000442325.2 ENST00000423854.2 ENST00000350494.6 ENST00000542824.1 |
ELP2
|
elongator acetyltransferase complex subunit 2 |
| chr15_-_81616446 | 0.03 |
ENST00000302824.6
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr7_+_13141010 | 0.03 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr2_-_231989808 | 0.03 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr8_+_120885949 | 0.03 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr12_+_76653682 | 0.03 |
ENST00000553247.1
|
RP11-54A9.1
|
RP11-54A9.1 |
| chr21_+_38792602 | 0.03 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr13_-_20110902 | 0.03 |
ENST00000390680.2
ENST00000382977.4 ENST00000382975.4 ENST00000457266.2 |
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr3_-_127541194 | 0.02 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr3_+_12392971 | 0.02 |
ENST00000287820.6
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr6_+_27925019 | 0.02 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr9_-_3489428 | 0.02 |
ENST00000451859.1
|
RFX3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr4_+_113739244 | 0.02 |
ENST00000503271.1
ENST00000503423.1 ENST00000506722.1 |
ANK2
|
ankyrin 2, neuronal |
| chr7_-_111424462 | 0.02 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr15_+_65204075 | 0.02 |
ENST00000380230.3
ENST00000357698.3 ENST00000395720.1 ENST00000496660.1 ENST00000319580.8 |
ANKDD1A
|
ankyrin repeat and death domain containing 1A |
| chr9_+_42704004 | 0.02 |
ENST00000457288.1
|
CBWD7
|
COBW domain containing 7 |
| chr8_+_125463048 | 0.02 |
ENST00000328599.3
|
TRMT12
|
tRNA methyltransferase 12 homolog (S. cerevisiae) |
| chr12_+_53848549 | 0.02 |
ENST00000439930.3
ENST00000548933.1 ENST00000562264.1 |
PCBP2
|
poly(rC) binding protein 2 |
| chr2_+_228735763 | 0.02 |
ENST00000373666.2
|
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr1_+_53308398 | 0.02 |
ENST00000371528.1
|
ZYG11A
|
zyg-11 family member A, cell cycle regulator |
| chr7_+_871559 | 0.02 |
ENST00000421580.1
|
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr1_-_89488510 | 0.02 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chr4_+_56212270 | 0.02 |
ENST00000264228.4
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr15_+_66585555 | 0.02 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr19_-_44388116 | 0.02 |
ENST00000587539.1
|
ZNF404
|
zinc finger protein 404 |
| chr8_-_108510224 | 0.02 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr15_+_25200108 | 0.02 |
ENST00000577949.1
ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF
SNRPN
|
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr6_+_106988986 | 0.02 |
ENST00000457437.1
ENST00000535438.1 |
AIM1
|
absent in melanoma 1 |
| chr1_-_85870177 | 0.02 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr13_+_24844819 | 0.02 |
ENST00000399949.2
|
SPATA13
|
spermatogenesis associated 13 |
| chr16_+_19467772 | 0.02 |
ENST00000219821.5
ENST00000561503.1 ENST00000564959.1 |
TMC5
|
transmembrane channel-like 5 |
| chr1_-_115124257 | 0.02 |
ENST00000369541.3
|
BCAS2
|
breast carcinoma amplified sequence 2 |
| chr17_-_42327236 | 0.02 |
ENST00000399246.2
|
AC003102.1
|
AC003102.1 |
| chr11_+_31531291 | 0.02 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chrX_+_15767971 | 0.02 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr19_-_44952635 | 0.02 |
ENST00000592308.1
ENST00000588931.1 ENST00000291187.4 |
ZNF229
|
zinc finger protein 229 |
| chr1_-_21377383 | 0.02 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr10_+_95326416 | 0.02 |
ENST00000371481.4
ENST00000371483.4 ENST00000604414.1 |
FFAR4
|
free fatty acid receptor 4 |
| chr22_+_18721427 | 0.02 |
ENST00000342888.3
|
AC008132.1
|
Uncharacterized protein |
| chr8_-_67976509 | 0.02 |
ENST00000518747.1
|
COPS5
|
COP9 signalosome subunit 5 |
| chr2_+_102624977 | 0.02 |
ENST00000441002.1
|
IL1R2
|
interleukin 1 receptor, type II |
| chr18_+_22006580 | 0.02 |
ENST00000284202.4
|
IMPACT
|
impact RWD domain protein |
| chr4_+_26344754 | 0.02 |
ENST00000515573.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr3_+_158787041 | 0.02 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr12_+_76653611 | 0.02 |
ENST00000550380.1
|
RP11-54A9.1
|
RP11-54A9.1 |
| chr8_+_27947746 | 0.02 |
ENST00000521015.1
ENST00000521570.1 |
ELP3
|
elongator acetyltransferase complex subunit 3 |
| chr17_-_18430160 | 0.02 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
| chr3_+_35681728 | 0.01 |
ENST00000421492.1
ENST00000458225.1 ENST00000337271.5 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr7_-_41742697 | 0.01 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr3_+_111758553 | 0.01 |
ENST00000419127.1
|
TMPRSS7
|
transmembrane protease, serine 7 |
| chr3_-_124606502 | 0.01 |
ENST00000483168.1
|
ITGB5
|
integrin, beta 5 |
| chr12_+_58335360 | 0.01 |
ENST00000300145.3
|
XRCC6BP1
|
XRCC6 binding protein 1 |
| chr13_-_20080080 | 0.01 |
ENST00000400103.2
|
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr17_+_75447326 | 0.01 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr17_+_58018269 | 0.01 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr1_+_113010056 | 0.01 |
ENST00000369686.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr14_-_95624227 | 0.01 |
ENST00000526495.1
|
DICER1
|
dicer 1, ribonuclease type III |
| chrX_-_135962923 | 0.01 |
ENST00000565438.1
|
RBMX
|
RNA binding motif protein, X-linked |
| chr3_+_138066539 | 0.01 |
ENST00000289104.4
|
MRAS
|
muscle RAS oncogene homolog |
| chr12_+_106751436 | 0.01 |
ENST00000228347.4
|
POLR3B
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr15_+_25200074 | 0.01 |
ENST00000390687.4
ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr1_-_201438282 | 0.01 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr20_+_43990576 | 0.01 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr16_-_58585513 | 0.01 |
ENST00000245138.4
ENST00000567285.1 |
CNOT1
|
CCR4-NOT transcription complex, subunit 1 |
| chr8_-_101724989 | 0.01 |
ENST00000517403.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_+_63989004 | 0.01 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr3_-_3221358 | 0.01 |
ENST00000424814.1
ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN
|
cereblon |
| chr11_+_114310237 | 0.01 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chr9_-_4666421 | 0.01 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr7_-_87856280 | 0.01 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chr12_-_120315074 | 0.01 |
ENST00000261833.7
ENST00000392521.2 |
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr14_+_96722539 | 0.01 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr1_+_155579979 | 0.01 |
ENST00000452804.2
ENST00000538143.1 ENST00000245564.2 ENST00000368341.4 |
MSTO1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr7_-_87856303 | 0.01 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr15_-_44116873 | 0.01 |
ENST00000267812.3
|
MFAP1
|
microfibrillar-associated protein 1 |
| chr1_+_22778337 | 0.01 |
ENST00000404138.1
ENST00000400239.2 ENST00000375647.4 ENST00000374651.4 |
ZBTB40
|
zinc finger and BTB domain containing 40 |
| chr7_+_133615169 | 0.01 |
ENST00000541309.1
|
EXOC4
|
exocyst complex component 4 |
| chr8_+_22225041 | 0.01 |
ENST00000289952.5
ENST00000524285.1 |
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chrX_-_15333736 | 0.01 |
ENST00000380470.3
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr19_+_50691437 | 0.01 |
ENST00000598205.1
|
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr14_-_74769759 | 0.01 |
ENST00000356924.4
|
ABCD4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr17_+_41561317 | 0.01 |
ENST00000540306.1
ENST00000262415.3 ENST00000605777.1 |
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr8_+_95558771 | 0.01 |
ENST00000391679.1
|
AC023632.1
|
HCG2009141; PRO2397; Uncharacterized protein |
| chr6_-_117747015 | 0.01 |
ENST00000368508.3
ENST00000368507.3 |
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase |
| chr4_+_41361616 | 0.01 |
ENST00000513024.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr17_-_6524159 | 0.01 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr8_+_27631903 | 0.01 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr3_+_155838337 | 0.01 |
ENST00000490337.1
ENST00000389636.5 |
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr12_-_25102252 | 0.01 |
ENST00000261192.7
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr3_+_160473996 | 0.01 |
ENST00000498165.1
|
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr17_+_22022437 | 0.01 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr13_+_96085847 | 0.01 |
ENST00000376873.3
|
CLDN10
|
claudin 10 |
| chr3_-_50378235 | 0.01 |
ENST00000357043.2
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr19_-_38743878 | 0.01 |
ENST00000587515.1
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr9_-_125667618 | 0.01 |
ENST00000423239.2
|
RC3H2
|
ring finger and CCCH-type domains 2 |
| chr12_-_58329888 | 0.01 |
ENST00000546580.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr5_+_67576109 | 0.01 |
ENST00000522084.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr10_+_96698406 | 0.01 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr19_-_2740036 | 0.01 |
ENST00000586572.1
ENST00000269740.4 |
AC006538.4
SLC39A3
|
Uncharacterized protein solute carrier family 39 (zinc transporter), member 3 |
| chr1_+_196743943 | 0.01 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr3_-_12883026 | 0.01 |
ENST00000396953.2
ENST00000457131.1 ENST00000435983.1 ENST00000273223.6 ENST00000396957.1 ENST00000429711.2 |
RPL32
|
ribosomal protein L32 |
| chr7_+_66386204 | 0.01 |
ENST00000341567.4
ENST00000607045.1 |
TMEM248
|
transmembrane protein 248 |
| chr8_-_17555164 | 0.01 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr13_+_97928395 | 0.01 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr9_+_71819927 | 0.01 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
| chr18_-_28622774 | 0.01 |
ENST00000434452.1
|
DSC3
|
desmocollin 3 |
| chr3_-_27498235 | 0.01 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr1_+_145525015 | 0.01 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chrX_-_19988382 | 0.01 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr2_+_210517895 | 0.01 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr2_-_38056538 | 0.01 |
ENST00000413792.1
|
LINC00211
|
long intergenic non-protein coding RNA 211 |
| chr11_+_22688150 | 0.01 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr4_+_70146217 | 0.01 |
ENST00000335568.5
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chr5_+_54455946 | 0.01 |
ENST00000503787.1
ENST00000296734.6 ENST00000515370.1 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr20_-_3762087 | 0.01 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr16_+_87636474 | 0.01 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr18_+_616672 | 0.00 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr1_+_196743912 | 0.00 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr1_+_202317815 | 0.00 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chrX_+_12809463 | 0.00 |
ENST00000380663.3
ENST00000380668.5 ENST00000398491.2 ENST00000489404.1 |
PRPS2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr1_-_21377447 | 0.00 |
ENST00000374937.3
ENST00000264211.8 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr3_-_72897545 | 0.00 |
ENST00000325599.8
|
SHQ1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr2_-_206950996 | 0.00 |
ENST00000414320.1
|
INO80D
|
INO80 complex subunit D |
| chr5_+_169011033 | 0.00 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr11_+_34999328 | 0.00 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr19_-_40596828 | 0.00 |
ENST00000414720.2
ENST00000455521.1 ENST00000340963.5 ENST00000595773.1 |
ZNF780A
|
zinc finger protein 780A |
| chr5_-_177207634 | 0.00 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chr12_-_123728548 | 0.00 |
ENST00000545406.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr16_-_90128733 | 0.00 |
ENST00000325921.6
|
PRDM7
|
PR domain containing 7 |
| chr5_+_169010638 | 0.00 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr10_-_51958906 | 0.00 |
ENST00000489640.1
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr6_-_154677900 | 0.00 |
ENST00000265198.4
ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr3_+_111630451 | 0.00 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr17_-_29641084 | 0.00 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr9_-_37465396 | 0.00 |
ENST00000307750.4
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr2_+_201173667 | 0.00 |
ENST00000409755.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr18_+_52258390 | 0.00 |
ENST00000321600.1
|
DYNAP
|
dynactin associated protein |
| chr7_+_13141097 | 0.00 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr18_-_32924372 | 0.00 |
ENST00000261332.6
ENST00000399061.3 |
ZNF24
|
zinc finger protein 24 |
| chrX_+_108779004 | 0.00 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr1_-_244006528 | 0.00 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr1_+_97188188 | 0.00 |
ENST00000541987.1
|
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr3_-_168865522 | 0.00 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr7_-_55583740 | 0.00 |
ENST00000453256.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.0 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) all-trans retinal binding(GO:0005503) |