Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
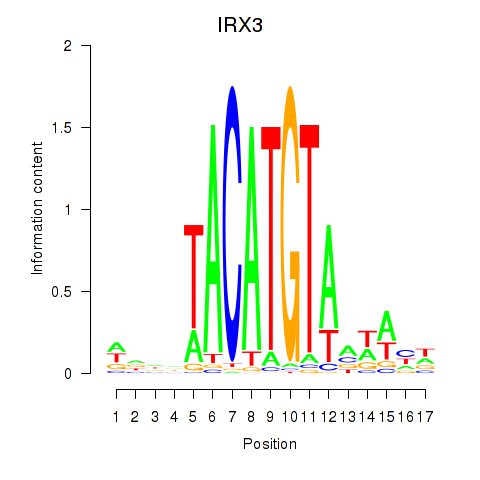
Results for IRX3
Z-value: 1.72
Transcription factors associated with IRX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX3
|
ENSG00000177508.11 | iroquois homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX3 | hg19_v2_chr16_-_54320675_54320715 | -0.58 | 4.2e-01 | Click! |
Activity profile of IRX3 motif
Sorted Z-values of IRX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_150866779 | 0.95 |
ENST00000370353.3
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr12_-_21757774 | 0.92 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver) |
| chr3_-_185641681 | 0.80 |
ENST00000259043.7
|
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr17_-_15469590 | 0.73 |
ENST00000312127.2
|
CDRT1
|
CMT duplicated region transcript 1; Uncharacterized protein |
| chr16_-_66864806 | 0.68 |
ENST00000566336.1
ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr5_-_156390230 | 0.67 |
ENST00000407087.3
ENST00000274532.2 |
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr14_+_39944025 | 0.66 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr22_+_22556057 | 0.65 |
ENST00000390286.2
|
IGLV11-55
|
immunoglobulin lambda variable 11-55 (non-functional) |
| chr5_+_81601166 | 0.64 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr7_+_40174565 | 0.62 |
ENST00000309930.5
ENST00000401647.2 ENST00000335693.4 ENST00000413931.1 ENST00000416370.1 ENST00000540834.1 |
C7orf10
|
succinylCoA:glutarate-CoA transferase |
| chr19_-_38743878 | 0.61 |
ENST00000587515.1
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr3_-_93747425 | 0.60 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr5_+_169011033 | 0.59 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr12_+_11187087 | 0.58 |
ENST00000601123.1
|
AC018630.1
|
Uncharacterized protein |
| chr5_-_156362666 | 0.57 |
ENST00000406964.1
|
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr13_-_44735393 | 0.55 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr5_+_136070614 | 0.54 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr3_-_48659193 | 0.53 |
ENST00000330862.3
|
TMEM89
|
transmembrane protein 89 |
| chr7_-_151330218 | 0.53 |
ENST00000476632.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr19_+_54960790 | 0.50 |
ENST00000443957.1
|
LENG8
|
leukocyte receptor cluster (LRC) member 8 |
| chr3_+_171561127 | 0.50 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr11_-_46113756 | 0.50 |
ENST00000531959.1
|
PHF21A
|
PHD finger protein 21A |
| chr12_+_2912363 | 0.49 |
ENST00000544366.1
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr17_-_34207295 | 0.49 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr8_+_52730143 | 0.47 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr9_+_135937365 | 0.47 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chr6_+_31691121 | 0.46 |
ENST00000480039.1
ENST00000375810.4 ENST00000375805.2 ENST00000375809.3 ENST00000375804.2 ENST00000375814.3 ENST00000375806.2 |
C6orf25
|
chromosome 6 open reading frame 25 |
| chr4_-_104119528 | 0.45 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr11_+_35201826 | 0.45 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr8_-_56986768 | 0.45 |
ENST00000523936.1
|
RPS20
|
ribosomal protein S20 |
| chr14_-_92247032 | 0.45 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr10_-_18944123 | 0.44 |
ENST00000606425.1
|
RP11-139J15.7
|
Uncharacterized protein |
| chr10_-_5227096 | 0.44 |
ENST00000488756.1
ENST00000334314.3 |
AKR1CL1
|
aldo-keto reductase family 1, member C-like 1 |
| chr5_+_56205081 | 0.44 |
ENST00000285947.2
ENST00000541720.1 |
SETD9
|
SET domain containing 9 |
| chr4_-_39367949 | 0.44 |
ENST00000503784.1
ENST00000349703.2 ENST00000381897.1 |
RFC1
|
replication factor C (activator 1) 1, 145kDa |
| chr20_+_10199566 | 0.43 |
ENST00000430336.1
|
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chrX_+_108779004 | 0.43 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr4_-_104119488 | 0.43 |
ENST00000514974.1
|
CENPE
|
centromere protein E, 312kDa |
| chr11_+_33037652 | 0.43 |
ENST00000311388.3
|
DEPDC7
|
DEP domain containing 7 |
| chr1_-_197744763 | 0.42 |
ENST00000422998.1
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr15_+_66585555 | 0.42 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr12_-_91546926 | 0.42 |
ENST00000550758.1
|
DCN
|
decorin |
| chr4_+_56212505 | 0.42 |
ENST00000505210.1
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr16_+_81070792 | 0.42 |
ENST00000564241.1
ENST00000565237.1 |
ATMIN
|
ATM interactor |
| chr8_-_56987061 | 0.42 |
ENST00000009589.3
ENST00000524349.1 ENST00000519606.1 ENST00000519807.1 ENST00000521262.1 ENST00000520627.1 |
RPS20
|
ribosomal protein S20 |
| chr15_+_72947079 | 0.42 |
ENST00000421285.3
|
GOLGA6B
|
golgin A6 family, member B |
| chr7_+_120591170 | 0.41 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family, member 3 |
| chr15_+_49170083 | 0.41 |
ENST00000530028.2
|
EID1
|
EP300 interacting inhibitor of differentiation 1 |
| chr14_+_89060739 | 0.41 |
ENST00000318308.6
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr1_+_145525015 | 0.41 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr1_+_154909803 | 0.40 |
ENST00000604546.1
|
RP11-307C12.13
|
RP11-307C12.13 |
| chr11_+_60739249 | 0.40 |
ENST00000542157.1
ENST00000433107.2 ENST00000452451.2 ENST00000352009.5 |
CD6
|
CD6 molecule |
| chr6_-_138833630 | 0.40 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr7_-_101212244 | 0.38 |
ENST00000451953.1
ENST00000434537.1 ENST00000437900.1 |
LINC01007
|
long intergenic non-protein coding RNA 1007 |
| chr17_-_26127525 | 0.37 |
ENST00000313735.6
|
NOS2
|
nitric oxide synthase 2, inducible |
| chr19_+_20959098 | 0.36 |
ENST00000360204.5
ENST00000594534.1 |
ZNF66
|
zinc finger protein 66 |
| chr10_-_112678904 | 0.36 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr4_-_144826682 | 0.36 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr16_-_21442874 | 0.36 |
ENST00000534903.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr13_+_37581115 | 0.36 |
ENST00000481013.1
|
EXOSC8
|
exosome component 8 |
| chr9_+_67968793 | 0.36 |
ENST00000417488.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr16_-_3493528 | 0.35 |
ENST00000301744.4
|
ZNF597
|
zinc finger protein 597 |
| chr16_-_9030515 | 0.35 |
ENST00000535863.1
ENST00000381886.4 |
USP7
|
ubiquitin specific peptidase 7 (herpes virus-associated) |
| chr12_-_123728548 | 0.35 |
ENST00000545406.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chrX_-_100129320 | 0.35 |
ENST00000372966.3
|
NOX1
|
NADPH oxidase 1 |
| chr12_-_13248598 | 0.35 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chrX_-_55187531 | 0.35 |
ENST00000489298.1
ENST00000477847.2 |
FAM104B
|
family with sequence similarity 104, member B |
| chr11_+_74303612 | 0.35 |
ENST00000527458.1
ENST00000532497.1 ENST00000530511.1 |
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr11_+_810221 | 0.35 |
ENST00000530398.1
|
RPLP2
|
ribosomal protein, large, P2 |
| chr4_+_74606223 | 0.35 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr19_+_58193337 | 0.35 |
ENST00000601064.1
ENST00000282296.5 ENST00000356715.4 |
ZNF551
|
zinc finger protein 551 |
| chr19_+_58193388 | 0.35 |
ENST00000596085.1
ENST00000594684.1 |
ZNF551
AC003006.7
|
zinc finger protein 551 Uncharacterized protein |
| chr12_-_100486668 | 0.35 |
ENST00000550544.1
ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L
|
UHRF1 binding protein 1-like |
| chr11_-_134123142 | 0.34 |
ENST00000392595.2
ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1
|
thymocyte nuclear protein 1 |
| chr10_+_52094298 | 0.34 |
ENST00000595931.1
|
AC069547.1
|
HCG1745369; PRO3073; Uncharacterized protein |
| chr4_-_186125077 | 0.34 |
ENST00000458385.2
ENST00000514798.1 ENST00000296775.6 |
KIAA1430
|
KIAA1430 |
| chr1_-_248005254 | 0.34 |
ENST00000355784.2
|
OR11L1
|
olfactory receptor, family 11, subfamily L, member 1 |
| chr10_+_13628933 | 0.33 |
ENST00000417658.1
ENST00000320054.4 |
PRPF18
|
pre-mRNA processing factor 18 |
| chr14_+_104177607 | 0.33 |
ENST00000429169.1
|
AL049840.1
|
Uncharacterized protein; cDNA FLJ53535 |
| chr4_-_84035905 | 0.33 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr4_+_76871883 | 0.33 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr8_+_101162812 | 0.33 |
ENST00000353107.3
ENST00000522439.1 |
POLR2K
|
polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa |
| chr10_+_90660832 | 0.33 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr4_-_38784592 | 0.32 |
ENST00000502321.1
|
TLR10
|
toll-like receptor 10 |
| chr10_+_79793518 | 0.32 |
ENST00000440692.1
ENST00000435275.1 ENST00000372360.3 ENST00000360830.4 |
RPS24
|
ribosomal protein S24 |
| chr5_+_54455946 | 0.32 |
ENST00000503787.1
ENST00000296734.6 ENST00000515370.1 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr4_+_130017268 | 0.32 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr4_+_75480629 | 0.32 |
ENST00000380846.3
|
AREGB
|
amphiregulin B |
| chr1_-_85156417 | 0.32 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr3_-_100566492 | 0.32 |
ENST00000528490.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr5_-_1551802 | 0.32 |
ENST00000503113.1
|
CTD-2245E15.3
|
CTD-2245E15.3 |
| chr6_-_27440837 | 0.32 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chrX_-_15353629 | 0.32 |
ENST00000333590.4
ENST00000428964.1 ENST00000542278.1 |
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr10_+_91461337 | 0.32 |
ENST00000260753.4
ENST00000416354.1 ENST00000394289.2 ENST00000371728.3 |
KIF20B
|
kinesin family member 20B |
| chr17_+_3379284 | 0.31 |
ENST00000263080.2
|
ASPA
|
aspartoacylase |
| chr8_+_94767072 | 0.31 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr12_+_21590549 | 0.31 |
ENST00000545178.1
ENST00000240651.9 |
PYROXD1
|
pyridine nucleotide-disulphide oxidoreductase domain 1 |
| chr15_+_35270552 | 0.31 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr17_+_22022437 | 0.31 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr17_+_58018269 | 0.30 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr7_-_35013217 | 0.30 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr11_+_17281900 | 0.30 |
ENST00000530527.1
|
NUCB2
|
nucleobindin 2 |
| chr14_+_56127960 | 0.30 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr4_-_103997862 | 0.30 |
ENST00000394785.3
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr10_+_91092241 | 0.30 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr21_+_25801041 | 0.30 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr10_-_112678976 | 0.30 |
ENST00000448814.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr3_-_27498235 | 0.29 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr15_-_56757329 | 0.29 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr11_+_12308447 | 0.29 |
ENST00000256186.2
|
MICALCL
|
MICAL C-terminal like |
| chr5_+_162887556 | 0.29 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr17_+_25958174 | 0.29 |
ENST00000313648.6
ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9
|
lectin, galactoside-binding, soluble, 9 |
| chr13_-_111522162 | 0.29 |
ENST00000538077.1
|
LINC00346
|
long intergenic non-protein coding RNA 346 |
| chr6_+_3982909 | 0.29 |
ENST00000356722.3
|
C6ORF50
|
C6ORF50 |
| chr15_+_69706643 | 0.29 |
ENST00000352331.4
ENST00000260363.4 |
KIF23
|
kinesin family member 23 |
| chr6_+_126221034 | 0.28 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr10_+_127371798 | 0.28 |
ENST00000596068.1
ENST00000607914.1 ENST00000415305.2 ENST00000449693.1 |
RP11-383C5.3
|
RP11-383C5.3 |
| chr13_-_88323218 | 0.28 |
ENST00000436290.2
ENST00000453832.2 ENST00000606590.1 |
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr4_-_75695366 | 0.28 |
ENST00000512743.1
|
BTC
|
betacellulin |
| chr6_-_153323801 | 0.28 |
ENST00000367233.5
ENST00000367231.5 ENST00000367230.1 |
MTRF1L
|
mitochondrial translational release factor 1-like |
| chr6_+_37012607 | 0.28 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr1_+_87458692 | 0.28 |
ENST00000370548.2
ENST00000356813.4 |
RP5-1052I5.2
HS2ST1
|
Heparan sulfate 2-O-sulfotransferase 1 heparan sulfate 2-O-sulfotransferase 1 |
| chr1_-_33116128 | 0.28 |
ENST00000436661.1
ENST00000373501.2 ENST00000341885.5 ENST00000468695.1 |
ZBTB8OS
|
zinc finger and BTB domain containing 8 opposite strand |
| chr7_-_91764108 | 0.28 |
ENST00000450723.1
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1 |
| chr2_-_38056538 | 0.27 |
ENST00000413792.1
|
LINC00211
|
long intergenic non-protein coding RNA 211 |
| chr11_+_86085778 | 0.27 |
ENST00000354755.1
ENST00000278487.3 ENST00000531271.1 ENST00000445632.2 |
CCDC81
|
coiled-coil domain containing 81 |
| chr14_+_89060749 | 0.27 |
ENST00000555900.1
ENST00000406216.3 ENST00000557737.1 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr7_-_130597935 | 0.27 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr9_-_135230336 | 0.27 |
ENST00000224140.5
ENST00000372169.2 ENST00000393220.1 |
SETX
|
senataxin |
| chr6_+_30908747 | 0.27 |
ENST00000462446.1
ENST00000304311.2 |
DPCR1
|
diffuse panbronchiolitis critical region 1 |
| chrX_+_49969405 | 0.27 |
ENST00000376042.1
|
CCNB3
|
cyclin B3 |
| chr2_+_169658928 | 0.27 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr2_-_201753980 | 0.26 |
ENST00000443398.1
ENST00000286175.8 ENST00000409449.1 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr2_+_202125219 | 0.26 |
ENST00000323492.7
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr5_+_68485433 | 0.26 |
ENST00000502689.1
|
CENPH
|
centromere protein H |
| chr10_-_35379524 | 0.26 |
ENST00000374751.3
ENST00000374742.1 ENST00000602371.1 |
CUL2
|
cullin 2 |
| chr12_+_58335360 | 0.26 |
ENST00000300145.3
|
XRCC6BP1
|
XRCC6 binding protein 1 |
| chr8_+_62737875 | 0.26 |
ENST00000523042.1
ENST00000518593.1 ENST00000519452.1 ENST00000519967.1 |
RP11-705O24.1
|
RP11-705O24.1 |
| chr12_+_79371565 | 0.25 |
ENST00000551304.1
|
SYT1
|
synaptotagmin I |
| chr11_+_57365150 | 0.25 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr10_-_61122220 | 0.25 |
ENST00000422313.2
ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C
|
family with sequence similarity 13, member C |
| chr5_+_159848854 | 0.25 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr19_-_38085633 | 0.25 |
ENST00000593133.1
ENST00000590751.1 ENST00000358744.3 ENST00000328550.2 ENST00000451802.2 |
ZNF571
|
zinc finger protein 571 |
| chr15_+_81225699 | 0.25 |
ENST00000560027.1
|
KIAA1199
|
KIAA1199 |
| chr2_-_231090344 | 0.24 |
ENST00000540870.1
ENST00000416610.1 |
SP110
|
SP110 nuclear body protein |
| chr10_-_14614311 | 0.24 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr2_+_109204909 | 0.24 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr11_-_74408739 | 0.24 |
ENST00000529912.1
|
CHRDL2
|
chordin-like 2 |
| chr19_+_41281282 | 0.24 |
ENST00000263369.3
|
MIA
|
melanoma inhibitory activity |
| chr2_-_88125471 | 0.24 |
ENST00000398146.3
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr19_+_53761545 | 0.24 |
ENST00000341702.3
|
VN1R2
|
vomeronasal 1 receptor 2 |
| chr3_+_57882061 | 0.24 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chr8_-_101962777 | 0.24 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr17_+_20978854 | 0.24 |
ENST00000456235.1
|
AC087393.1
|
AC087393.1 |
| chr13_-_95248511 | 0.24 |
ENST00000261296.5
|
TGDS
|
TDP-glucose 4,6-dehydratase |
| chr19_+_7580103 | 0.24 |
ENST00000596712.1
|
ZNF358
|
zinc finger protein 358 |
| chr19_-_23456996 | 0.24 |
ENST00000594653.1
|
RP11-15H20.7
|
RP11-15H20.7 |
| chr10_-_96829246 | 0.23 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr6_+_153552455 | 0.23 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr16_+_19535235 | 0.23 |
ENST00000565376.2
ENST00000396208.2 |
CCP110
|
centriolar coiled coil protein 110kDa |
| chr7_-_16685422 | 0.23 |
ENST00000306999.2
|
ANKMY2
|
ankyrin repeat and MYND domain containing 2 |
| chr4_+_78829479 | 0.23 |
ENST00000504901.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr17_-_8066843 | 0.23 |
ENST00000404970.3
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2) |
| chr5_+_112073544 | 0.23 |
ENST00000257430.4
ENST00000508376.2 |
APC
|
adenomatous polyposis coli |
| chr17_+_75450099 | 0.23 |
ENST00000586433.1
|
SEPT9
|
septin 9 |
| chr18_+_21529811 | 0.23 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr10_-_14050522 | 0.23 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr16_+_84209539 | 0.23 |
ENST00000569735.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr16_+_19535133 | 0.23 |
ENST00000396212.2
ENST00000381396.5 |
CCP110
|
centriolar coiled coil protein 110kDa |
| chr10_-_30638090 | 0.23 |
ENST00000421701.1
ENST00000263063.4 |
MTPAP
|
mitochondrial poly(A) polymerase |
| chr4_-_103998439 | 0.23 |
ENST00000503230.1
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr5_+_41904431 | 0.23 |
ENST00000381647.2
|
C5orf51
|
chromosome 5 open reading frame 51 |
| chr3_-_57583052 | 0.22 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr12_-_26986076 | 0.22 |
ENST00000381340.3
|
ITPR2
|
inositol 1,4,5-trisphosphate receptor, type 2 |
| chr5_+_96079240 | 0.22 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr14_+_56127989 | 0.22 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr3_+_14061155 | 0.22 |
ENST00000532924.1
|
TPRXL
|
tetra-peptide repeat homeobox-like |
| chr12_-_133464118 | 0.22 |
ENST00000540963.1
|
CHFR
|
checkpoint with forkhead and ring finger domains, E3 ubiquitin protein ligase |
| chr15_+_69706585 | 0.22 |
ENST00000559279.1
ENST00000395392.2 |
KIF23
|
kinesin family member 23 |
| chr9_-_21305312 | 0.22 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr6_-_75828774 | 0.22 |
ENST00000493109.2
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr9_+_4679555 | 0.22 |
ENST00000381858.1
ENST00000381854.3 |
CDC37L1
|
cell division cycle 37-like 1 |
| chr2_-_26251481 | 0.22 |
ENST00000599234.1
|
AC013449.1
|
Uncharacterized protein |
| chr4_+_79567057 | 0.22 |
ENST00000503259.1
ENST00000507802.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr10_+_105881779 | 0.22 |
ENST00000369729.3
|
SFR1
|
SWI5-dependent recombination repair 1 |
| chr1_+_227751231 | 0.22 |
ENST00000343776.5
ENST00000608949.1 ENST00000397097.3 |
ZNF678
|
zinc finger protein 678 |
| chr4_+_71600144 | 0.22 |
ENST00000502653.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr3_-_12883026 | 0.22 |
ENST00000396953.2
ENST00000457131.1 ENST00000435983.1 ENST00000273223.6 ENST00000396957.1 ENST00000429711.2 |
RPL32
|
ribosomal protein L32 |
| chr4_+_189321881 | 0.22 |
ENST00000512839.1
ENST00000513313.1 |
LINC01060
|
long intergenic non-protein coding RNA 1060 |
| chr22_+_22673051 | 0.22 |
ENST00000390289.2
|
IGLV5-52
|
immunoglobulin lambda variable 5-52 |
| chr5_-_86708833 | 0.21 |
ENST00000256897.4
|
CCNH
|
cyclin H |
| chr11_-_14358620 | 0.21 |
ENST00000531421.1
|
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr8_+_132952112 | 0.21 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr1_+_240408560 | 0.21 |
ENST00000441342.1
ENST00000545751.1 |
FMN2
|
formin 2 |
| chr11_-_104905840 | 0.21 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr16_+_67840986 | 0.21 |
ENST00000561639.1
ENST00000567852.1 ENST00000565148.1 ENST00000388833.3 ENST00000561654.1 ENST00000431934.2 |
TSNAXIP1
|
translin-associated factor X interacting protein 1 |
| chr16_+_22518495 | 0.21 |
ENST00000541154.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr10_-_14614095 | 0.21 |
ENST00000482277.1
ENST00000378462.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr2_+_182850743 | 0.21 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr2_+_46926048 | 0.21 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 | 0.5 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.1 | 0.6 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.5 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.5 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.9 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.5 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.3 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 0.3 | GO:2000562 | natural killer cell tolerance induction(GO:0002519) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.3 | GO:0033488 | cholesterol biosynthetic process via 24,25-dihydrolanosterol(GO:0033488) |
| 0.1 | 0.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.4 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.7 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.3 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.5 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 0.6 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.2 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.1 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.3 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.1 | 0.7 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 0.4 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.4 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.2 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.2 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.2 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.2 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.8 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0043308 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.2 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0019254 | amino-acid betaine catabolic process(GO:0006579) carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.6 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:0032618 | interleukin-15 production(GO:0032618) |
| 0.0 | 0.1 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.7 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.6 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.2 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.5 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.8 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.4 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.2 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.9 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 | 0.3 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.2 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.2 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.3 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.2 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 0.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.1 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0003051 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0048023 | mammillary body development(GO:0021767) positive regulation of melanin biosynthetic process(GO:0048023) mammillary axonal complex development(GO:0061373) positive regulation of secondary metabolite biosynthetic process(GO:1900378) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.6 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.5 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.2 | GO:0051957 | positive regulation of amino acid transport(GO:0051957) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.1 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 0.3 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.1 | GO:2000230 | regulation of cholesterol transporter activity(GO:0060694) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0035966 | response to unfolded protein(GO:0006986) response to topologically incorrect protein(GO:0035966) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.2 | GO:2000674 | regulation of type B pancreatic cell apoptotic process(GO:2000674) |
| 0.0 | 0.3 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.2 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.2 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) |
| 0.0 | 0.1 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.2 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.0 | GO:0070487 | monocyte aggregation(GO:0070487) |
| 0.0 | 0.4 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.0 | 0.0 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.5 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.1 | 0.8 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 1.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.5 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.6 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.2 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.1 | 1.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.2 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.1 | 0.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.2 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 0.2 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.2 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.3 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.3 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.6 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0097526 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.2 | 1.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.4 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.1 | 0.5 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.3 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.4 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 0.6 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.4 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.3 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.3 | GO:0008398 | sterol 14-demethylase activity(GO:0008398) |
| 0.1 | 0.5 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.3 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 1.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.3 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 0.6 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.7 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.2 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) tetrahydrobiopterin binding(GO:0034617) |
| 0.1 | 0.5 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.2 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.2 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.3 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.3 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 1.6 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 0.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 1.3 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0047291 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0050308 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.0 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.3 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.0 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.2 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |