Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
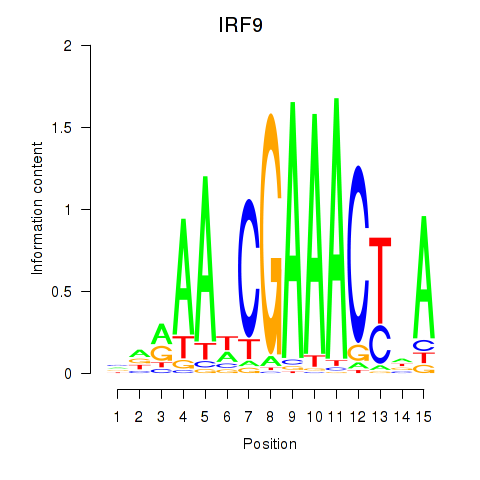
Results for IRF9
Z-value: 0.96
Transcription factors associated with IRF9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF9
|
ENSG00000213928.4 | interferon regulatory factor 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF9 | hg19_v2_chr14_+_24630465_24630531 | -0.83 | 1.7e-01 | Click! |
Activity profile of IRF9 motif
Sorted Z-values of IRF9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_6659153 | 0.65 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr12_-_121477039 | 0.60 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr12_-_121476959 | 0.53 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr12_-_121476750 | 0.42 |
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr16_+_53412368 | 0.37 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr2_-_163175133 | 0.36 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr19_+_16254488 | 0.36 |
ENST00000588246.1
ENST00000593031.1 |
HSH2D
|
hematopoietic SH2 domain containing |
| chr10_+_91061712 | 0.27 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr10_+_91092241 | 0.27 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr17_-_40264692 | 0.27 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr4_-_76944621 | 0.26 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr4_-_147442817 | 0.26 |
ENST00000507030.1
|
SLC10A7
|
solute carrier family 10, member 7 |
| chr9_+_33264861 | 0.24 |
ENST00000223500.8
|
CHMP5
|
charged multivesicular body protein 5 |
| chr9_-_32526184 | 0.24 |
ENST00000545044.1
ENST00000379868.1 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr3_+_159706537 | 0.24 |
ENST00000305579.2
ENST00000480787.1 ENST00000466512.1 |
IL12A
|
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
| chr9_+_33265011 | 0.23 |
ENST00000419016.2
|
CHMP5
|
charged multivesicular body protein 5 |
| chr4_+_186990298 | 0.23 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr16_-_74734672 | 0.22 |
ENST00000306247.7
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr1_-_154167124 | 0.22 |
ENST00000515609.1
|
TPM3
|
tropomyosin 3 |
| chr9_+_74764278 | 0.21 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr2_-_7005785 | 0.21 |
ENST00000256722.5
ENST00000404168.1 ENST00000458098.1 |
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr12_+_113344755 | 0.20 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr17_+_46908350 | 0.20 |
ENST00000258947.3
ENST00000509507.1 ENST00000448105.2 ENST00000570513.1 ENST00000509415.1 ENST00000513119.1 ENST00000416445.2 ENST00000508679.1 ENST00000505071.1 |
CALCOCO2
|
calcium binding and coiled-coil domain 2 |
| chr4_-_75719896 | 0.20 |
ENST00000395743.3
|
BTC
|
betacellulin |
| chr2_-_152118276 | 0.20 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr4_-_147442982 | 0.19 |
ENST00000511374.1
ENST00000264986.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr1_+_79115503 | 0.19 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr14_+_94577074 | 0.18 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr3_-_179169181 | 0.18 |
ENST00000497513.1
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr9_+_70971815 | 0.16 |
ENST00000396392.1
ENST00000396396.1 |
PGM5
|
phosphoglucomutase 5 |
| chr10_+_91174486 | 0.16 |
ENST00000416601.1
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5 |
| chr2_+_33701684 | 0.15 |
ENST00000442390.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr2_-_55920952 | 0.15 |
ENST00000447944.2
|
PNPT1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr2_-_37384175 | 0.14 |
ENST00000411537.2
ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr3_-_146262365 | 0.14 |
ENST00000448787.2
|
PLSCR1
|
phospholipid scramblase 1 |
| chr4_+_89378261 | 0.14 |
ENST00000264350.3
|
HERC5
|
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr17_-_74489215 | 0.14 |
ENST00000585701.1
ENST00000591192.1 ENST00000589526.1 |
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila) |
| chr3_-_28389922 | 0.14 |
ENST00000415852.1
|
AZI2
|
5-azacytidine induced 2 |
| chr3_-_146262352 | 0.14 |
ENST00000462666.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr3_-_28390120 | 0.14 |
ENST00000334100.6
|
AZI2
|
5-azacytidine induced 2 |
| chr12_+_113344582 | 0.14 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr2_+_33701707 | 0.14 |
ENST00000425210.1
ENST00000444784.1 ENST00000423159.1 |
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr19_-_17516449 | 0.13 |
ENST00000252593.6
|
BST2
|
bone marrow stromal cell antigen 2 |
| chr7_-_105332084 | 0.13 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr3_-_122283424 | 0.13 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr3_-_28390581 | 0.12 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr17_+_6659354 | 0.12 |
ENST00000574907.1
|
XAF1
|
XIAP associated factor 1 |
| chr15_+_45003675 | 0.12 |
ENST00000558401.1
ENST00000559916.1 ENST00000544417.1 |
B2M
|
beta-2-microglobulin |
| chr13_+_50070491 | 0.12 |
ENST00000496612.1
ENST00000357596.3 ENST00000485919.1 ENST00000442195.1 |
PHF11
|
PHD finger protein 11 |
| chr3_-_141747439 | 0.12 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr4_-_100140331 | 0.11 |
ENST00000407820.2
ENST00000394897.1 ENST00000508558.1 ENST00000394899.2 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr3_-_142166846 | 0.11 |
ENST00000463916.1
ENST00000544157.1 |
XRN1
|
5'-3' exoribonuclease 1 |
| chr3_-_122283079 | 0.11 |
ENST00000471785.1
ENST00000466126.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr14_-_67981870 | 0.11 |
ENST00000555994.1
|
TMEM229B
|
transmembrane protein 229B |
| chr6_-_8102714 | 0.11 |
ENST00000502429.1
ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr9_+_74764340 | 0.11 |
ENST00000376986.1
ENST00000358399.3 |
GDA
|
guanine deaminase |
| chr10_+_91174314 | 0.11 |
ENST00000371795.4
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5 |
| chr9_-_32526299 | 0.11 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr4_+_37892682 | 0.11 |
ENST00000508802.1
ENST00000261439.4 ENST00000402522.1 |
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr3_-_142166796 | 0.10 |
ENST00000392981.2
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr17_+_41363854 | 0.10 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr4_-_169401628 | 0.10 |
ENST00000514748.1
ENST00000512371.1 ENST00000260184.7 ENST00000505890.1 ENST00000511577.1 |
DDX60L
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
| chr5_-_142783694 | 0.10 |
ENST00000394466.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr3_-_28390415 | 0.10 |
ENST00000414162.1
ENST00000420543.2 |
AZI2
|
5-azacytidine induced 2 |
| chr1_-_150738261 | 0.09 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr6_-_105584560 | 0.09 |
ENST00000336775.5
|
BVES
|
blood vessel epicardial substance |
| chr3_+_122399444 | 0.09 |
ENST00000474629.2
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr4_-_77134742 | 0.09 |
ENST00000452464.2
|
SCARB2
|
scavenger receptor class B, member 2 |
| chr3_-_28390298 | 0.09 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr4_+_146402997 | 0.09 |
ENST00000512019.1
|
SMAD1
|
SMAD family member 1 |
| chr5_-_95297534 | 0.09 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr2_-_152146385 | 0.08 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr3_-_27764190 | 0.08 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr3_-_141747459 | 0.08 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr11_-_4414880 | 0.08 |
ENST00000254436.7
ENST00000543625.1 |
TRIM21
|
tripartite motif containing 21 |
| chr5_-_137911049 | 0.08 |
ENST00000297185.3
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin) |
| chr4_+_146402925 | 0.08 |
ENST00000302085.4
|
SMAD1
|
SMAD family member 1 |
| chr1_+_66820058 | 0.08 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr3_-_121379739 | 0.07 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr11_-_60720002 | 0.07 |
ENST00000538739.1
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr1_+_162602244 | 0.07 |
ENST00000367922.3
ENST00000367921.3 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr9_+_102668915 | 0.07 |
ENST00000259400.6
ENST00000531035.1 ENST00000525640.1 ENST00000534052.1 ENST00000526607.1 |
STX17
|
syntaxin 17 |
| chr12_+_113344811 | 0.07 |
ENST00000551241.1
ENST00000553185.1 ENST00000550689.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr4_-_169239921 | 0.07 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr2_-_106810783 | 0.07 |
ENST00000283148.7
|
UXS1
|
UDP-glucuronate decarboxylase 1 |
| chr5_-_142784003 | 0.06 |
ENST00000416954.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr1_+_59486129 | 0.06 |
ENST00000438195.1
ENST00000424308.1 |
RP4-794H19.4
|
RP4-794H19.4 |
| chr21_+_42798124 | 0.06 |
ENST00000417963.1
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr21_+_42798094 | 0.06 |
ENST00000398598.3
ENST00000455164.2 ENST00000424365.1 |
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chrX_-_39956656 | 0.06 |
ENST00000397354.3
ENST00000378444.4 |
BCOR
|
BCL6 corepressor |
| chr2_-_106810742 | 0.06 |
ENST00000409501.3
ENST00000428048.2 ENST00000441952.1 ENST00000457835.1 ENST00000540130.1 |
UXS1
|
UDP-glucuronate decarboxylase 1 |
| chr19_+_10222189 | 0.05 |
ENST00000321826.4
|
P2RY11
|
purinergic receptor P2Y, G-protein coupled, 11 |
| chr13_+_50070077 | 0.05 |
ENST00000378319.3
ENST00000426879.1 |
PHF11
|
PHD finger protein 11 |
| chr2_+_7073174 | 0.05 |
ENST00000416587.1
|
RNF144A
|
ring finger protein 144A |
| chr2_+_205410516 | 0.05 |
ENST00000406610.2
ENST00000462231.1 |
PARD3B
|
par-3 family cell polarity regulator beta |
| chr11_-_57335280 | 0.05 |
ENST00000287156.4
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr5_-_95297678 | 0.05 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr6_-_82462425 | 0.05 |
ENST00000369754.3
ENST00000320172.6 ENST00000369756.3 |
FAM46A
|
family with sequence similarity 46, member A |
| chr1_+_158979792 | 0.05 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr16_-_15736881 | 0.05 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chrX_-_48858667 | 0.05 |
ENST00000376423.4
ENST00000376441.1 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr14_-_67981916 | 0.05 |
ENST00000357461.2
|
TMEM229B
|
transmembrane protein 229B |
| chr10_-_25241499 | 0.04 |
ENST00000376378.1
ENST00000376376.3 ENST00000320152.6 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr11_-_615570 | 0.04 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chrX_-_48858630 | 0.04 |
ENST00000376425.3
ENST00000376444.3 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr11_-_57334732 | 0.04 |
ENST00000526659.1
ENST00000527022.1 |
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr17_-_54991369 | 0.04 |
ENST00000537230.1
|
TRIM25
|
tripartite motif containing 25 |
| chr11_-_86383157 | 0.04 |
ENST00000393324.3
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr14_+_100531615 | 0.04 |
ENST00000392920.3
|
EVL
|
Enah/Vasp-like |
| chr3_-_146262428 | 0.03 |
ENST00000486631.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr10_-_91174215 | 0.03 |
ENST00000371837.1
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase |
| chr9_-_33264557 | 0.03 |
ENST00000473781.1
ENST00000488499.1 |
BAG1
|
BCL2-associated athanogene |
| chr20_+_11898507 | 0.03 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr1_+_158979686 | 0.03 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr5_-_59481406 | 0.03 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr3_-_146262637 | 0.03 |
ENST00000472349.1
ENST00000342435.4 |
PLSCR1
|
phospholipid scramblase 1 |
| chr11_+_5646213 | 0.03 |
ENST00000429814.2
|
TRIM34
|
tripartite motif containing 34 |
| chr3_-_122283100 | 0.03 |
ENST00000492382.1
ENST00000462315.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr18_+_3262954 | 0.03 |
ENST00000584539.1
|
MYL12B
|
myosin, light chain 12B, regulatory |
| chr10_+_115439699 | 0.03 |
ENST00000369315.1
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr6_-_31324943 | 0.03 |
ENST00000412585.2
ENST00000434333.1 |
HLA-B
|
major histocompatibility complex, class I, B |
| chr15_+_91643442 | 0.02 |
ENST00000394232.1
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr4_-_77135046 | 0.02 |
ENST00000264896.2
|
SCARB2
|
scavenger receptor class B, member 2 |
| chr17_+_78234625 | 0.02 |
ENST00000508628.2
ENST00000582970.1 ENST00000456466.1 ENST00000319921.4 |
RNF213
|
ring finger protein 213 |
| chr2_+_205410723 | 0.02 |
ENST00000358768.2
ENST00000351153.1 ENST00000349953.3 |
PARD3B
|
par-3 family cell polarity regulator beta |
| chr1_-_234667504 | 0.02 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr10_+_115439282 | 0.02 |
ENST00000369321.2
ENST00000345633.4 |
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chrX_-_63005405 | 0.02 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr6_+_126240442 | 0.02 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr15_+_58702742 | 0.02 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr15_+_74287118 | 0.02 |
ENST00000563500.1
|
PML
|
promyelocytic leukemia |
| chr3_+_122283064 | 0.02 |
ENST00000296161.4
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr3_+_151986709 | 0.02 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr10_+_115439630 | 0.02 |
ENST00000369318.3
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr21_+_42797958 | 0.02 |
ENST00000419044.1
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr10_-_95242044 | 0.01 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr5_-_142783175 | 0.01 |
ENST00000231509.3
ENST00000394464.2 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr17_+_6658878 | 0.01 |
ENST00000574394.1
|
XAF1
|
XIAP associated factor 1 |
| chr20_-_56195449 | 0.01 |
ENST00000541799.1
|
ZBP1
|
Z-DNA binding protein 1 |
| chr1_+_236687881 | 0.01 |
ENST00000526634.1
|
LGALS8
|
lectin, galactoside-binding, soluble, 8 |
| chr10_-_95241951 | 0.01 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr18_-_67624412 | 0.00 |
ENST00000580335.1
|
CD226
|
CD226 molecule |
| chr3_+_122283175 | 0.00 |
ENST00000383661.3
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr19_+_10196981 | 0.00 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.3 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.1 | 0.6 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.2 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:2000627 | rRNA import into mitochondrion(GO:0035928) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.0 | 0.1 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) |
| 0.0 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:0033319 | UDP-D-xylose metabolic process(GO:0033319) UDP-D-xylose biosynthetic process(GO:0033320) |
| 0.0 | 0.1 | GO:1904432 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.0 | 0.5 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.4 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.2 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.1 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.0 | 0.5 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.3 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.8 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 1.7 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.1 | GO:0050709 | negative regulation of protein secretion(GO:0050709) |
| 0.0 | 0.0 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.3 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.1 | 0.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0048040 | UDP-glucuronate decarboxylase activity(GO:0048040) |
| 0.0 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 1.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.7 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 2.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |