Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for IRF3
Z-value: 0.89
Transcription factors associated with IRF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF3
|
ENSG00000126456.11 | interferon regulatory factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF3 | hg19_v2_chr19_-_50169064_50169132 | 0.74 | 2.6e-01 | Click! |
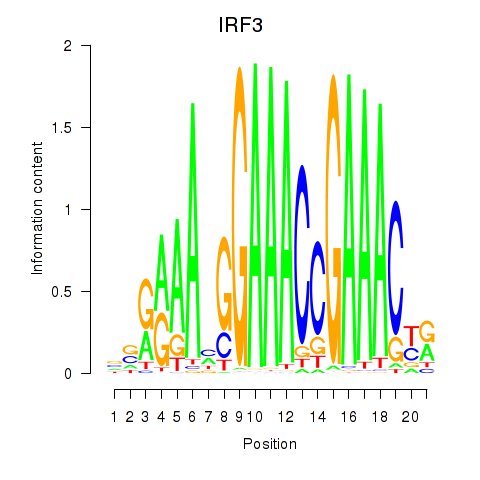
Activity profile of IRF3 motif
Sorted Z-values of IRF3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_28223229 | 0.89 |
ENST00000566073.1
|
XPO6
|
exportin 6 |
| chr9_-_14322319 | 0.76 |
ENST00000606230.1
|
NFIB
|
nuclear factor I/B |
| chr12_+_113416265 | 0.53 |
ENST00000449768.2
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr12_+_113416340 | 0.50 |
ENST00000552756.1
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr1_+_948803 | 0.47 |
ENST00000379389.4
|
ISG15
|
ISG15 ubiquitin-like modifier |
| chr12_+_113416191 | 0.44 |
ENST00000342315.4
ENST00000392583.2 |
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr3_-_172241250 | 0.41 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr15_-_55611306 | 0.39 |
ENST00000563262.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr19_-_17516449 | 0.36 |
ENST00000252593.6
|
BST2
|
bone marrow stromal cell antigen 2 |
| chr3_+_122399697 | 0.34 |
ENST00000494811.1
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr4_-_169239921 | 0.30 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr1_-_146644036 | 0.29 |
ENST00000425272.2
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr5_-_60139347 | 0.29 |
ENST00000511799.1
|
ELOVL7
|
ELOVL fatty acid elongase 7 |
| chr19_+_39926791 | 0.27 |
ENST00000594990.1
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr2_-_163175133 | 0.27 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr16_-_11350036 | 0.27 |
ENST00000332029.2
|
SOCS1
|
suppressor of cytokine signaling 1 |
| chr19_-_47735918 | 0.26 |
ENST00000449228.1
ENST00000300880.7 ENST00000341983.4 |
BBC3
|
BCL2 binding component 3 |
| chr12_+_113376249 | 0.26 |
ENST00000551007.1
ENST00000548514.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr1_+_110577229 | 0.25 |
ENST00000369795.3
ENST00000369794.2 |
STRIP1
|
striatin interacting protein 1 |
| chr20_+_61436146 | 0.25 |
ENST00000290291.6
|
OGFR
|
opioid growth factor receptor |
| chr7_+_142829162 | 0.24 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr7_-_92777606 | 0.23 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr2_+_201981527 | 0.23 |
ENST00000441224.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr4_-_156875003 | 0.23 |
ENST00000433477.3
|
CTSO
|
cathepsin O |
| chr9_-_90589586 | 0.22 |
ENST00000325303.8
ENST00000375883.3 |
CDK20
|
cyclin-dependent kinase 20 |
| chr1_+_59486129 | 0.22 |
ENST00000438195.1
ENST00000424308.1 |
RP4-794H19.4
|
RP4-794H19.4 |
| chr11_-_615942 | 0.22 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr1_+_210502238 | 0.21 |
ENST00000545154.1
ENST00000537898.1 ENST00000391905.3 ENST00000545781.1 ENST00000261458.3 ENST00000308852.6 |
HHAT
|
hedgehog acyltransferase |
| chr11_-_57334732 | 0.21 |
ENST00000526659.1
ENST00000527022.1 |
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr8_+_109455830 | 0.21 |
ENST00000524143.1
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr1_-_161015663 | 0.20 |
ENST00000534633.1
|
USF1
|
upstream transcription factor 1 |
| chr16_-_67970990 | 0.19 |
ENST00000358514.4
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr20_+_388791 | 0.19 |
ENST00000441733.1
ENST00000353660.3 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr19_+_10196981 | 0.19 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr11_-_615570 | 0.18 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr21_+_42792442 | 0.18 |
ENST00000398600.2
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr22_-_36635598 | 0.18 |
ENST00000454728.1
|
APOL2
|
apolipoprotein L, 2 |
| chr17_-_61517572 | 0.17 |
ENST00000582997.1
|
CYB561
|
cytochrome b561 |
| chr1_+_79115503 | 0.17 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr7_+_100728720 | 0.17 |
ENST00000306085.6
ENST00000412507.1 |
TRIM56
|
tripartite motif containing 56 |
| chr15_+_55611401 | 0.16 |
ENST00000566999.1
|
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr8_+_22446763 | 0.16 |
ENST00000450780.2
ENST00000430850.2 ENST00000447849.1 |
AC037459.4
|
Uncharacterized protein |
| chr22_+_36649056 | 0.16 |
ENST00000397278.3
ENST00000422706.1 ENST00000426053.1 ENST00000319136.4 |
APOL1
|
apolipoprotein L, 1 |
| chr7_-_139756791 | 0.16 |
ENST00000489809.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr10_-_22292675 | 0.16 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr15_-_88799948 | 0.15 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr12_+_6561190 | 0.15 |
ENST00000544021.1
ENST00000266556.7 |
TAPBPL
|
TAP binding protein-like |
| chr19_+_50431959 | 0.15 |
ENST00000595125.1
|
ATF5
|
activating transcription factor 5 |
| chr6_+_32821924 | 0.15 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr22_-_36635225 | 0.15 |
ENST00000529194.1
|
APOL2
|
apolipoprotein L, 2 |
| chr14_+_35451880 | 0.15 |
ENST00000554803.1
ENST00000555746.1 |
SRP54
|
signal recognition particle 54kDa |
| chr6_+_26087509 | 0.14 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr6_+_26087646 | 0.13 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr4_-_169401628 | 0.13 |
ENST00000514748.1
ENST00000512371.1 ENST00000260184.7 ENST00000505890.1 ENST00000511577.1 |
DDX60L
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
| chr17_+_6659153 | 0.13 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr3_-_53079281 | 0.13 |
ENST00000394750.1
|
SFMBT1
|
Scm-like with four mbt domains 1 |
| chr2_-_231989808 | 0.12 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr11_+_66008698 | 0.12 |
ENST00000531597.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr5_-_95158375 | 0.12 |
ENST00000512469.2
ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX
|
glutaredoxin (thioltransferase) |
| chr3_-_125802765 | 0.12 |
ENST00000514891.1
ENST00000512470.1 ENST00000504035.1 ENST00000360370.4 ENST00000513723.1 ENST00000510651.1 ENST00000514333.1 |
SLC41A3
|
solute carrier family 41, member 3 |
| chr6_+_26402517 | 0.12 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr1_+_236557569 | 0.12 |
ENST00000334232.4
|
EDARADD
|
EDAR-associated death domain |
| chr10_+_91087651 | 0.12 |
ENST00000371818.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr3_-_125802998 | 0.11 |
ENST00000514677.1
|
SLC41A3
|
solute carrier family 41, member 3 |
| chr12_+_14369524 | 0.11 |
ENST00000538329.1
|
RP11-134N1.2
|
RP11-134N1.2 |
| chr2_+_201980961 | 0.11 |
ENST00000342795.5
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr17_+_36861735 | 0.11 |
ENST00000378137.5
ENST00000325718.7 |
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr17_+_21730180 | 0.10 |
ENST00000584398.1
|
UBBP4
|
ubiquitin B pseudogene 4 |
| chr19_+_50432400 | 0.10 |
ENST00000423777.2
ENST00000600336.1 ENST00000597227.1 |
ATF5
|
activating transcription factor 5 |
| chr2_-_190044480 | 0.10 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr20_-_25038804 | 0.10 |
ENST00000323482.4
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1 |
| chr15_+_64428529 | 0.10 |
ENST00000560861.1
|
SNX1
|
sorting nexin 1 |
| chr12_+_133614062 | 0.09 |
ENST00000540031.1
ENST00000536123.1 |
ZNF84
|
zinc finger protein 84 |
| chr9_-_90589402 | 0.09 |
ENST00000375871.4
ENST00000605159.1 ENST00000336654.5 |
CDK20
|
cyclin-dependent kinase 20 |
| chr6_+_26440700 | 0.09 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chrX_+_154610428 | 0.09 |
ENST00000354514.4
|
H2AFB2
|
H2A histone family, member B2 |
| chr20_+_388679 | 0.09 |
ENST00000356286.5
ENST00000475269.1 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr17_+_21729899 | 0.09 |
ENST00000583708.1
|
UBBP4
|
ubiquitin B pseudogene 4 |
| chrX_+_10031499 | 0.09 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr2_+_239756671 | 0.08 |
ENST00000448943.2
|
TWIST2
|
twist family bHLH transcription factor 2 |
| chr11_-_796197 | 0.08 |
ENST00000530360.1
ENST00000528606.1 ENST00000320230.5 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr1_-_177939041 | 0.08 |
ENST00000308284.6
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr12_-_10022735 | 0.08 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr12_+_97300995 | 0.08 |
ENST00000266742.4
ENST00000429527.2 ENST00000554226.1 ENST00000557478.1 ENST00000557092.1 ENST00000411739.2 ENST00000553609.1 |
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr16_+_66968343 | 0.08 |
ENST00000417689.1
ENST00000561697.1 ENST00000317091.4 ENST00000566182.1 |
CES2
|
carboxylesterase 2 |
| chr11_+_124735282 | 0.07 |
ENST00000397801.1
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr2_+_99771527 | 0.07 |
ENST00000415142.1
ENST00000436234.1 |
LIPT1
|
lipoyltransferase 1 |
| chr12_-_54582655 | 0.07 |
ENST00000504338.1
ENST00000514685.1 ENST00000504797.1 ENST00000513838.1 ENST00000505128.1 ENST00000337581.3 ENST00000503306.1 ENST00000243112.5 ENST00000514196.1 ENST00000506169.1 ENST00000507904.1 ENST00000508394.2 |
SMUG1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| chr2_-_152118276 | 0.07 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr5_+_131746575 | 0.07 |
ENST00000337752.2
ENST00000378947.3 ENST00000407797.1 |
C5orf56
|
chromosome 5 open reading frame 56 |
| chr10_+_91092241 | 0.07 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr9_-_95640218 | 0.07 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr2_+_201980827 | 0.07 |
ENST00000309955.3
ENST00000443227.1 ENST00000341222.6 ENST00000355558.4 ENST00000340870.5 ENST00000341582.6 |
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr17_+_79373540 | 0.07 |
ENST00000307745.7
|
RP11-1055B8.7
|
BAH and coiled-coil domain-containing protein 1 |
| chr12_+_133613937 | 0.07 |
ENST00000539354.1
ENST00000542874.1 ENST00000438628.2 |
ZNF84
|
zinc finger protein 84 |
| chr3_+_63898275 | 0.07 |
ENST00000538065.1
|
ATXN7
|
ataxin 7 |
| chr1_+_25598872 | 0.06 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen |
| chr22_+_36044411 | 0.06 |
ENST00000409652.4
|
APOL6
|
apolipoprotein L, 6 |
| chr7_-_32931387 | 0.06 |
ENST00000304056.4
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr1_+_112298514 | 0.06 |
ENST00000536167.1
|
DDX20
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 |
| chr20_-_47894936 | 0.06 |
ENST00000371754.4
|
ZNFX1
|
zinc finger, NFX1-type containing 1 |
| chr2_-_119605253 | 0.06 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr17_+_8316442 | 0.06 |
ENST00000582812.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr12_-_133613794 | 0.06 |
ENST00000443154.3
|
RP11-386I8.6
|
RP11-386I8.6 |
| chr1_-_161014731 | 0.06 |
ENST00000368020.1
|
USF1
|
upstream transcription factor 1 |
| chr5_+_140071178 | 0.06 |
ENST00000508522.1
ENST00000448069.2 |
HARS2
|
histidyl-tRNA synthetase 2, mitochondrial |
| chr11_+_5646213 | 0.05 |
ENST00000429814.2
|
TRIM34
|
tripartite motif containing 34 |
| chr16_-_29875057 | 0.05 |
ENST00000219789.6
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr11_-_57335280 | 0.05 |
ENST00000287156.4
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr3_-_123339343 | 0.05 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr9_-_100954910 | 0.05 |
ENST00000375077.4
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr19_+_10196781 | 0.05 |
ENST00000253110.11
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr20_+_388935 | 0.05 |
ENST00000382181.2
ENST00000400247.3 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr4_+_87856129 | 0.05 |
ENST00000395146.4
ENST00000507468.1 |
AFF1
|
AF4/FMR2 family, member 1 |
| chr10_+_48355024 | 0.05 |
ENST00000395702.2
ENST00000442001.1 ENST00000433077.1 ENST00000436850.1 ENST00000494156.1 ENST00000586537.1 |
ZNF488
|
zinc finger protein 488 |
| chr19_+_50887585 | 0.04 |
ENST00000440232.2
ENST00000601098.1 ENST00000599857.1 ENST00000593887.1 |
POLD1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr5_-_77656175 | 0.04 |
ENST00000513755.1
ENST00000421004.3 |
CTD-2037K23.2
|
CTD-2037K23.2 |
| chr4_+_118955500 | 0.04 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr8_+_27238147 | 0.04 |
ENST00000412793.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr6_-_90529418 | 0.04 |
ENST00000439638.1
ENST00000369393.3 ENST00000428876.1 |
MDN1
|
MDN1, midasin homolog (yeast) |
| chr6_+_41748500 | 0.04 |
ENST00000458694.1
ENST00000359201.5 |
PRICKLE4
|
prickle homolog 4 (Drosophila) |
| chr19_+_49977466 | 0.04 |
ENST00000596435.1
ENST00000344019.3 ENST00000597551.1 ENST00000204637.2 ENST00000600429.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr14_+_73525144 | 0.04 |
ENST00000261973.7
ENST00000540173.1 |
RBM25
|
RNA binding motif protein 25 |
| chr3_-_71114066 | 0.04 |
ENST00000485326.2
|
FOXP1
|
forkhead box P1 |
| chr4_+_124320665 | 0.03 |
ENST00000394339.2
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr1_+_151129103 | 0.03 |
ENST00000368910.3
|
TNFAIP8L2
|
tumor necrosis factor, alpha-induced protein 8-like 2 |
| chr20_+_20033158 | 0.03 |
ENST00000340348.6
ENST00000377309.2 ENST00000389656.3 ENST00000377306.1 ENST00000245957.5 ENST00000377303.2 ENST00000475466.1 |
C20orf26
|
chromosome 20 open reading frame 26 |
| chrX_+_57313113 | 0.03 |
ENST00000374900.4
|
FAAH2
|
fatty acid amide hydrolase 2 |
| chr20_-_33872518 | 0.03 |
ENST00000374436.3
|
EIF6
|
eukaryotic translation initiation factor 6 |
| chr15_-_62352570 | 0.03 |
ENST00000261517.5
ENST00000395896.4 ENST00000395898.3 |
VPS13C
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr2_+_99771418 | 0.03 |
ENST00000393473.2
ENST00000393477.3 ENST00000393474.3 ENST00000340066.1 ENST00000393471.2 ENST00000449211.1 ENST00000434566.1 ENST00000410042.1 |
LIPT1
MRPL30
|
lipoyltransferase 1 39S ribosomal protein L30, mitochondrial |
| chr20_-_61847455 | 0.03 |
ENST00000370334.4
|
YTHDF1
|
YTH domain family, member 1 |
| chr1_-_177939348 | 0.03 |
ENST00000464631.2
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr10_-_102046417 | 0.03 |
ENST00000370372.2
|
BLOC1S2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr1_-_161015752 | 0.03 |
ENST00000435396.1
ENST00000368021.3 |
USF1
|
upstream transcription factor 1 |
| chr9_+_140083099 | 0.03 |
ENST00000322310.5
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr1_+_200011711 | 0.03 |
ENST00000544748.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr10_+_115439630 | 0.03 |
ENST00000369318.3
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr17_+_6658878 | 0.03 |
ENST00000574394.1
|
XAF1
|
XIAP associated factor 1 |
| chr1_-_182558374 | 0.03 |
ENST00000367559.3
ENST00000539397.1 |
RNASEL
|
ribonuclease L (2',5'-oligoisoadenylate synthetase-dependent) |
| chr14_-_70883708 | 0.02 |
ENST00000256366.4
|
SYNJ2BP
|
synaptojanin 2 binding protein |
| chr4_+_130017268 | 0.02 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr3_+_63897605 | 0.02 |
ENST00000487717.1
|
ATXN7
|
ataxin 7 |
| chr14_+_73525229 | 0.02 |
ENST00000527432.1
ENST00000531500.1 ENST00000525321.1 ENST00000526754.1 |
RBM25
|
RNA binding motif protein 25 |
| chr17_-_7166500 | 0.02 |
ENST00000575313.1
ENST00000397317.4 |
CLDN7
|
claudin 7 |
| chr3_-_125803105 | 0.02 |
ENST00000346785.5
ENST00000315891.6 |
SLC41A3
|
solute carrier family 41, member 3 |
| chr7_-_37382360 | 0.02 |
ENST00000455119.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr22_+_18632666 | 0.02 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr9_-_140082983 | 0.02 |
ENST00000323927.2
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chr11_-_60720002 | 0.02 |
ENST00000538739.1
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr2_-_231084820 | 0.02 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr5_+_140749803 | 0.02 |
ENST00000576222.1
|
PCDHGB3
|
protocadherin gamma subfamily B, 3 |
| chr3_+_122399444 | 0.02 |
ENST00000474629.2
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr11_+_125461607 | 0.02 |
ENST00000527606.1
|
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr3_+_187957646 | 0.02 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr22_+_31892261 | 0.02 |
ENST00000432498.1
ENST00000540643.1 ENST00000443326.1 ENST00000414585.1 |
SFI1
|
Sfi1 homolog, spindle assembly associated (yeast) |
| chr6_+_42018614 | 0.02 |
ENST00000465926.1
ENST00000482432.1 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr14_-_75518129 | 0.02 |
ENST00000556257.1
ENST00000557648.1 ENST00000553263.1 ENST00000355774.2 ENST00000380968.2 ENST00000238662.7 |
MLH3
|
mutL homolog 3 |
| chr1_+_76190357 | 0.01 |
ENST00000370834.5
ENST00000541113.1 ENST00000543667.1 ENST00000420607.2 |
ACADM
|
acyl-CoA dehydrogenase, C-4 to C-12 straight chain |
| chr14_-_91976794 | 0.01 |
ENST00000555462.1
|
SMEK1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr14_-_24615523 | 0.01 |
ENST00000559056.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr6_-_41039567 | 0.01 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr6_-_110964453 | 0.01 |
ENST00000413605.2
|
CDK19
|
cyclin-dependent kinase 19 |
| chr6_-_32821599 | 0.01 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr5_-_180288248 | 0.01 |
ENST00000512132.1
ENST00000506439.1 ENST00000502412.1 ENST00000359141.6 |
ZFP62
|
ZFP62 zinc finger protein |
| chr22_+_36649170 | 0.01 |
ENST00000438034.1
ENST00000427990.1 ENST00000347595.7 ENST00000397279.4 ENST00000433768.1 ENST00000440669.2 |
APOL1
|
apolipoprotein L, 1 |
| chr22_+_39436862 | 0.01 |
ENST00000381565.2
ENST00000452957.2 |
APOBEC3F
APOBEC3G
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G |
| chr1_-_146644122 | 0.01 |
ENST00000254101.3
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr6_+_26402465 | 0.01 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr9_+_74764278 | 0.01 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr6_+_42018251 | 0.01 |
ENST00000372978.3
ENST00000494547.1 ENST00000456846.2 ENST00000372982.4 ENST00000472818.1 ENST00000372977.3 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr17_+_6659354 | 0.01 |
ENST00000574907.1
|
XAF1
|
XIAP associated factor 1 |
| chr2_-_231084659 | 0.01 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chr7_+_134832808 | 0.00 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr4_+_87928140 | 0.00 |
ENST00000307808.6
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr9_+_77112244 | 0.00 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr4_-_85771168 | 0.00 |
ENST00000514071.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr20_-_56195449 | 0.00 |
ENST00000541799.1
|
ZBP1
|
Z-DNA binding protein 1 |
| chr3_+_51977294 | 0.00 |
ENST00000498510.1
|
PARP3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr3_+_122283175 | 0.00 |
ENST00000383661.3
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr6_-_32806506 | 0.00 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr4_-_87855851 | 0.00 |
ENST00000473559.1
|
C4orf36
|
chromosome 4 open reading frame 36 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.1 | 1.5 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.1 | 0.4 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) |
| 0.1 | 0.4 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 0.8 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.4 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.2 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.3 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.0 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.3 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.3 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.1 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.2 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.3 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.7 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.3 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.4 | GO:0033093 | Weibel-Palade body(GO:0033093) melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.3 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 2.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |