Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
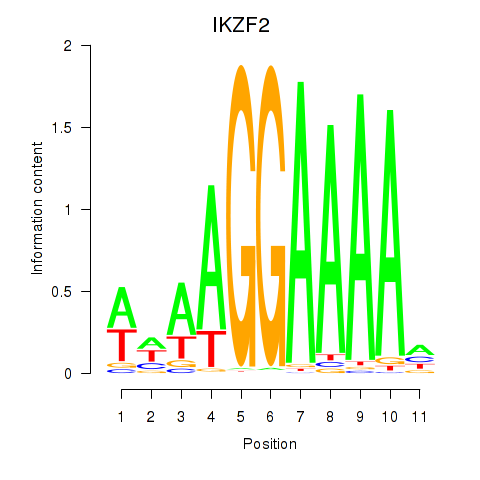
Results for IKZF2
Z-value: 0.76
Transcription factors associated with IKZF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IKZF2
|
ENSG00000030419.12 | IKAROS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IKZF2 | hg19_v2_chr2_-_214015111_214015179 | -0.86 | 1.4e-01 | Click! |
Activity profile of IKZF2 motif
Sorted Z-values of IKZF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_68845417 | 0.65 |
ENST00000542875.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr19_-_39735646 | 0.59 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr17_-_15469590 | 0.46 |
ENST00000312127.2
|
CDRT1
|
CMT duplicated region transcript 1; Uncharacterized protein |
| chr21_+_35107346 | 0.34 |
ENST00000456489.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr10_-_10504285 | 0.32 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr14_+_32546274 | 0.27 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr2_-_119605253 | 0.26 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr9_-_16705069 | 0.22 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr9_+_132099158 | 0.21 |
ENST00000444125.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr14_-_35591156 | 0.21 |
ENST00000554361.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr11_-_69867159 | 0.20 |
ENST00000528507.1
|
RP11-626H12.2
|
RP11-626H12.2 |
| chr1_+_40627038 | 0.20 |
ENST00000372771.4
|
RLF
|
rearranged L-myc fusion |
| chr3_+_193853927 | 0.20 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chrX_+_21874105 | 0.19 |
ENST00000429584.2
|
YY2
|
YY2 transcription factor |
| chr10_+_63422695 | 0.18 |
ENST00000330194.2
ENST00000389639.3 |
C10orf107
|
chromosome 10 open reading frame 107 |
| chr12_-_49581152 | 0.18 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr17_+_41150793 | 0.18 |
ENST00000586277.1
|
RPL27
|
ribosomal protein L27 |
| chrX_+_73164149 | 0.17 |
ENST00000602938.1
ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr5_-_175815565 | 0.17 |
ENST00000509257.1
ENST00000507413.1 ENST00000510123.1 |
NOP16
|
NOP16 nucleolar protein |
| chr12_-_76461249 | 0.17 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr6_-_97345689 | 0.17 |
ENST00000316149.7
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 |
| chr10_-_112678904 | 0.17 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr5_+_27472371 | 0.17 |
ENST00000512067.1
ENST00000510165.1 ENST00000505775.1 ENST00000514844.1 ENST00000503107.1 |
LINC01021
|
long intergenic non-protein coding RNA 1021 |
| chr12_-_110511424 | 0.17 |
ENST00000548191.1
|
C12orf76
|
chromosome 12 open reading frame 76 |
| chr11_-_19263145 | 0.17 |
ENST00000532666.1
ENST00000527884.1 |
E2F8
|
E2F transcription factor 8 |
| chr3_+_57882061 | 0.17 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chrX_-_15683147 | 0.16 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr12_-_109025849 | 0.16 |
ENST00000228463.6
|
SELPLG
|
selectin P ligand |
| chr8_+_97597148 | 0.16 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chrX_+_100663243 | 0.16 |
ENST00000316594.5
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr3_-_27764190 | 0.16 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr6_-_131384347 | 0.16 |
ENST00000530481.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr12_-_4488872 | 0.16 |
ENST00000237837.1
|
FGF23
|
fibroblast growth factor 23 |
| chr12_-_8693539 | 0.16 |
ENST00000299663.3
|
CLEC4E
|
C-type lectin domain family 4, member E |
| chr14_+_32546145 | 0.15 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr10_-_112678976 | 0.15 |
ENST00000448814.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr16_+_20911978 | 0.15 |
ENST00000562740.1
|
LYRM1
|
LYR motif containing 1 |
| chr17_-_26127525 | 0.15 |
ENST00000313735.6
|
NOS2
|
nitric oxide synthase 2, inducible |
| chr7_-_135194822 | 0.15 |
ENST00000428680.2
ENST00000315544.5 ENST00000423368.2 ENST00000451834.1 ENST00000361528.4 ENST00000356162.4 ENST00000541284.1 |
CNOT4
|
CCR4-NOT transcription complex, subunit 4 |
| chr18_+_19668021 | 0.15 |
ENST00000579830.1
|
RP11-595B24.2
|
Uncharacterized protein |
| chr5_-_34043310 | 0.15 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr2_-_45795145 | 0.14 |
ENST00000535761.1
|
SRBD1
|
S1 RNA binding domain 1 |
| chr1_+_24019099 | 0.14 |
ENST00000443624.1
ENST00000458455.1 |
RPL11
|
ribosomal protein L11 |
| chr12_-_68845165 | 0.14 |
ENST00000360485.3
ENST00000441255.2 |
RP11-81H14.2
|
RP11-81H14.2 |
| chr1_+_168148169 | 0.14 |
ENST00000367833.2
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr14_-_45431091 | 0.14 |
ENST00000579157.1
ENST00000396128.4 ENST00000556500.1 |
KLHL28
|
kelch-like family member 28 |
| chr14_+_56078695 | 0.14 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr16_-_28374829 | 0.14 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chr19_-_43709772 | 0.14 |
ENST00000596907.1
ENST00000451895.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr6_-_131384412 | 0.13 |
ENST00000445890.2
ENST00000368128.2 ENST00000337057.3 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr2_+_98330009 | 0.13 |
ENST00000264972.5
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa |
| chr11_-_8892464 | 0.13 |
ENST00000527347.1
ENST00000526241.1 ENST00000526126.1 ENST00000530938.1 ENST00000526057.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr10_-_112678692 | 0.13 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr6_-_18265050 | 0.13 |
ENST00000397239.3
|
DEK
|
DEK oncogene |
| chr3_+_178866199 | 0.13 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr17_-_48785216 | 0.13 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr20_-_50808236 | 0.13 |
ENST00000361387.2
|
ZFP64
|
ZFP64 zinc finger protein |
| chrX_+_135230712 | 0.13 |
ENST00000535737.1
|
FHL1
|
four and a half LIM domains 1 |
| chr5_+_122847908 | 0.13 |
ENST00000511130.2
ENST00000512718.3 |
CSNK1G3
|
casein kinase 1, gamma 3 |
| chr7_+_79765071 | 0.12 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr1_-_67600639 | 0.12 |
ENST00000544837.1
ENST00000603691.1 |
C1orf141
|
chromosome 1 open reading frame 141 |
| chr15_-_66545995 | 0.12 |
ENST00000395614.1
ENST00000288745.3 ENST00000422354.1 ENST00000395625.2 ENST00000360698.4 ENST00000409699.2 |
MEGF11
|
multiple EGF-like-domains 11 |
| chrX_+_80457442 | 0.12 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chr6_-_131384373 | 0.12 |
ENST00000392427.3
ENST00000525271.1 ENST00000527411.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chrX_+_73164167 | 0.12 |
ENST00000414209.1
ENST00000602895.1 ENST00000453317.1 ENST00000602546.1 ENST00000602985.1 ENST00000415215.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr1_+_10490441 | 0.12 |
ENST00000470413.2
ENST00000309048.3 |
APITD1-CORT
APITD1
|
APITD1-CORT readthrough apoptosis-inducing, TAF9-like domain 1 |
| chr4_-_134070250 | 0.12 |
ENST00000505289.1
ENST00000509715.1 |
RP11-9G1.3
|
RP11-9G1.3 |
| chr10_+_81892477 | 0.12 |
ENST00000372263.3
|
PLAC9
|
placenta-specific 9 |
| chr14_+_32476072 | 0.12 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chr14_+_64932210 | 0.12 |
ENST00000394718.4
|
AKAP5
|
A kinase (PRKA) anchor protein 5 |
| chr2_+_46926048 | 0.11 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr2_+_101437487 | 0.11 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr1_+_168148273 | 0.11 |
ENST00000367830.3
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr17_+_34848049 | 0.11 |
ENST00000588902.1
ENST00000591067.1 |
ZNHIT3
|
zinc finger, HIT-type containing 3 |
| chr4_-_157892498 | 0.11 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr1_+_150954493 | 0.11 |
ENST00000368947.4
|
ANXA9
|
annexin A9 |
| chr17_-_38938786 | 0.11 |
ENST00000301656.3
|
KRT27
|
keratin 27 |
| chrX_+_135252050 | 0.11 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr9_+_67968793 | 0.11 |
ENST00000417488.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr15_+_69706643 | 0.11 |
ENST00000352331.4
ENST00000260363.4 |
KIF23
|
kinesin family member 23 |
| chr15_+_43985725 | 0.11 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr5_+_159436120 | 0.11 |
ENST00000522793.1
ENST00000231238.5 |
TTC1
|
tetratricopeptide repeat domain 1 |
| chr14_+_89060739 | 0.11 |
ENST00000318308.6
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr20_-_50808290 | 0.11 |
ENST00000346617.4
ENST00000371515.4 ENST00000371518.2 |
ZFP64
|
ZFP64 zinc finger protein |
| chr6_-_110964453 | 0.11 |
ENST00000413605.2
|
CDK19
|
cyclin-dependent kinase 19 |
| chr2_+_232135245 | 0.11 |
ENST00000446447.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr1_-_242612779 | 0.11 |
ENST00000427495.1
|
PLD5
|
phospholipase D family, member 5 |
| chr8_-_10987745 | 0.11 |
ENST00000400102.3
|
AF131215.5
|
AF131215.5 |
| chr1_-_197169672 | 0.10 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr17_-_62499334 | 0.10 |
ENST00000579996.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr12_-_80328949 | 0.10 |
ENST00000450142.2
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A |
| chr12_-_74686314 | 0.10 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr5_+_122847781 | 0.10 |
ENST00000395412.1
ENST00000395411.1 ENST00000345990.4 |
CSNK1G3
|
casein kinase 1, gamma 3 |
| chr16_+_82660635 | 0.10 |
ENST00000567445.1
ENST00000446376.2 |
CDH13
|
cadherin 13 |
| chr1_-_11118896 | 0.10 |
ENST00000465788.1
|
SRM
|
spermidine synthase |
| chr1_-_242612726 | 0.10 |
ENST00000459864.1
|
PLD5
|
phospholipase D family, member 5 |
| chr7_-_151574191 | 0.10 |
ENST00000287878.4
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr18_-_24237339 | 0.10 |
ENST00000580191.1
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr4_+_130017268 | 0.10 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr7_-_64451414 | 0.10 |
ENST00000282869.6
|
ZNF117
|
zinc finger protein 117 |
| chr4_+_95174445 | 0.10 |
ENST00000509418.1
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr5_+_61874562 | 0.10 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr8_-_17942432 | 0.10 |
ENST00000381733.4
ENST00000314146.10 |
ASAH1
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
| chr4_-_157892055 | 0.10 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr20_+_57466357 | 0.10 |
ENST00000371095.3
ENST00000371085.3 ENST00000354359.7 ENST00000265620.7 |
GNAS
|
GNAS complex locus |
| chr3_+_171844762 | 0.10 |
ENST00000443501.1
|
FNDC3B
|
fibronectin type III domain containing 3B |
| chr2_+_136289030 | 0.10 |
ENST00000409478.1
ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1
|
R3H domain containing 1 |
| chr17_+_39382900 | 0.10 |
ENST00000377721.3
ENST00000455970.2 |
KRTAP9-2
|
keratin associated protein 9-2 |
| chr15_-_90358048 | 0.10 |
ENST00000300060.6
ENST00000560137.1 |
ANPEP
|
alanyl (membrane) aminopeptidase |
| chr14_+_45431379 | 0.09 |
ENST00000361577.3
ENST00000361462.2 ENST00000382233.2 |
FAM179B
|
family with sequence similarity 179, member B |
| chr2_-_136288740 | 0.09 |
ENST00000264159.6
ENST00000536680.1 |
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chrX_-_19689106 | 0.09 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr1_-_82726817 | 0.09 |
ENST00000420549.1
|
RP11-147G16.1
|
RP11-147G16.1 |
| chrX_-_102941596 | 0.09 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr12_+_79371565 | 0.09 |
ENST00000551304.1
|
SYT1
|
synaptotagmin I |
| chr20_-_50808525 | 0.09 |
ENST00000216923.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr6_+_32944119 | 0.09 |
ENST00000606059.1
|
BRD2
|
bromodomain containing 2 |
| chr2_+_187350883 | 0.09 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr10_+_112679301 | 0.09 |
ENST00000265277.5
ENST00000369452.4 |
SHOC2
|
soc-2 suppressor of clear homolog (C. elegans) |
| chr12_-_100656134 | 0.09 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr16_-_12184159 | 0.09 |
ENST00000312019.2
|
RP11-276H1.3
|
RP11-276H1.3 |
| chrX_+_135251835 | 0.09 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr3_+_156393349 | 0.09 |
ENST00000473702.1
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr12_-_6602955 | 0.09 |
ENST00000543703.1
|
MRPL51
|
mitochondrial ribosomal protein L51 |
| chr9_+_70856899 | 0.09 |
ENST00000377342.5
ENST00000478048.1 |
CBWD3
|
COBW domain containing 3 |
| chr8_-_133637624 | 0.09 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr3_-_100566492 | 0.09 |
ENST00000528490.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chrX_-_45629661 | 0.09 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr21_-_35284635 | 0.09 |
ENST00000429238.1
|
AP000304.12
|
AP000304.12 |
| chr5_-_96518907 | 0.09 |
ENST00000508447.1
ENST00000283109.3 |
RIOK2
|
RIO kinase 2 |
| chr20_-_14318248 | 0.09 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr4_-_185776854 | 0.09 |
ENST00000511703.1
|
RP11-701P16.5
|
RP11-701P16.5 |
| chr2_+_46926326 | 0.09 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chrX_+_135251783 | 0.09 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr12_-_80328905 | 0.09 |
ENST00000547330.1
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A |
| chr1_+_68150744 | 0.09 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr4_-_185303418 | 0.08 |
ENST00000610223.1
ENST00000608785.1 |
RP11-290F5.1
|
RP11-290F5.1 |
| chr1_+_111889212 | 0.08 |
ENST00000369737.4
|
PIFO
|
primary cilia formation |
| chr13_+_73629107 | 0.08 |
ENST00000539231.1
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr4_-_83765613 | 0.08 |
ENST00000503937.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr1_-_31666767 | 0.08 |
ENST00000530145.1
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr1_-_234831899 | 0.08 |
ENST00000442382.1
|
RP4-781K5.6
|
RP4-781K5.6 |
| chr13_-_73356009 | 0.08 |
ENST00000377780.4
ENST00000377767.4 |
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae) |
| chr1_+_104615595 | 0.08 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr1_-_47779762 | 0.08 |
ENST00000371877.3
ENST00000360380.3 ENST00000337817.5 ENST00000447475.2 |
STIL
|
SCL/TAL1 interrupting locus |
| chr11_-_108093329 | 0.08 |
ENST00000278612.8
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus |
| chrX_+_123097014 | 0.08 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr1_+_161719552 | 0.08 |
ENST00000367943.4
|
DUSP12
|
dual specificity phosphatase 12 |
| chr3_+_46618727 | 0.08 |
ENST00000296145.5
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr6_-_31846744 | 0.08 |
ENST00000414427.1
ENST00000229729.6 ENST00000375562.4 |
SLC44A4
|
solute carrier family 44, member 4 |
| chr6_-_11779403 | 0.08 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr5_-_43515231 | 0.08 |
ENST00000306862.2
|
C5orf34
|
chromosome 5 open reading frame 34 |
| chr3_+_97983088 | 0.08 |
ENST00000383696.2
|
OR5H6
|
olfactory receptor, family 5, subfamily H, member 6 (gene/pseudogene) |
| chr10_+_105036909 | 0.08 |
ENST00000369849.4
|
INA
|
internexin neuronal intermediate filament protein, alpha |
| chr9_+_2159850 | 0.08 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr6_+_149539767 | 0.08 |
ENST00000606202.1
ENST00000536230.1 ENST00000445901.1 |
TAB2
RP1-111D6.3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr15_+_69706585 | 0.08 |
ENST00000559279.1
ENST00000395392.2 |
KIF23
|
kinesin family member 23 |
| chr14_-_36988882 | 0.08 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr17_+_1182948 | 0.08 |
ENST00000333813.3
|
TUSC5
|
tumor suppressor candidate 5 |
| chr6_-_42185583 | 0.08 |
ENST00000053468.3
|
MRPS10
|
mitochondrial ribosomal protein S10 |
| chr20_-_61733657 | 0.08 |
ENST00000608031.1
ENST00000447910.2 |
HAR1B
|
highly accelerated region 1B (non-protein coding) |
| chr6_+_13925318 | 0.08 |
ENST00000423553.2
ENST00000537388.1 |
RNF182
|
ring finger protein 182 |
| chr13_+_20532848 | 0.08 |
ENST00000382874.2
|
ZMYM2
|
zinc finger, MYM-type 2 |
| chr19_-_43709703 | 0.08 |
ENST00000599391.1
ENST00000244295.9 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr18_-_21852143 | 0.08 |
ENST00000399443.3
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr9_-_73483926 | 0.08 |
ENST00000396283.1
ENST00000361823.5 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr12_-_323689 | 0.08 |
ENST00000428720.1
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr10_+_115511434 | 0.08 |
ENST00000369312.4
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr15_-_66679019 | 0.08 |
ENST00000568216.1
ENST00000562124.1 ENST00000570251.1 |
TIPIN
|
TIMELESS interacting protein |
| chr17_-_16118835 | 0.08 |
ENST00000582357.1
ENST00000436828.1 ENST00000411510.1 ENST00000268712.3 |
NCOR1
|
nuclear receptor corepressor 1 |
| chr13_+_97874574 | 0.08 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr1_-_193075180 | 0.08 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr16_+_72459838 | 0.07 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3 |
| chr20_-_43589109 | 0.07 |
ENST00000372813.3
|
TOMM34
|
translocase of outer mitochondrial membrane 34 |
| chr12_-_56367101 | 0.07 |
ENST00000549233.2
|
PMEL
|
premelanosome protein |
| chr20_-_45981138 | 0.07 |
ENST00000446994.2
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr3_-_71632894 | 0.07 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr4_+_39184024 | 0.07 |
ENST00000399820.3
ENST00000509560.1 ENST00000512112.1 ENST00000288634.7 ENST00000506503.1 |
WDR19
|
WD repeat domain 19 |
| chr14_+_63671105 | 0.07 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr2_-_197791441 | 0.07 |
ENST00000409475.1
ENST00000354764.4 ENST00000374738.3 |
PGAP1
|
post-GPI attachment to proteins 1 |
| chr15_-_83837983 | 0.07 |
ENST00000562702.1
|
HDGFRP3
|
Hepatoma-derived growth factor-related protein 3 |
| chr20_+_57209828 | 0.07 |
ENST00000598340.1
|
MGC4294
|
HCG1785561; MGC4294 protein; Uncharacterized protein |
| chr8_+_121457642 | 0.07 |
ENST00000305949.1
|
MTBP
|
Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa |
| chr12_+_69979113 | 0.07 |
ENST00000299300.6
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr11_-_46142948 | 0.07 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chrX_-_74743080 | 0.07 |
ENST00000373367.3
|
ZDHHC15
|
zinc finger, DHHC-type containing 15 |
| chr21_+_17909594 | 0.07 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_-_161337662 | 0.07 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr15_-_55541227 | 0.07 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr14_-_92506371 | 0.07 |
ENST00000267622.4
|
TRIP11
|
thyroid hormone receptor interactor 11 |
| chr15_+_63050785 | 0.07 |
ENST00000472902.1
|
TLN2
|
talin 2 |
| chr14_-_55493763 | 0.07 |
ENST00000455555.1
ENST00000360586.3 ENST00000421192.1 ENST00000420358.2 |
WDHD1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr4_+_72052964 | 0.07 |
ENST00000264485.5
ENST00000425175.1 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr9_-_127952187 | 0.07 |
ENST00000451402.1
ENST00000415905.1 |
PPP6C
|
protein phosphatase 6, catalytic subunit |
| chrX_+_129040122 | 0.07 |
ENST00000394422.3
ENST00000371051.5 |
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr8_-_101964265 | 0.07 |
ENST00000395958.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr14_+_64970427 | 0.07 |
ENST00000553583.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr1_+_158979680 | 0.07 |
ENST00000368131.4
ENST00000340979.6 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr6_+_126221034 | 0.07 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr19_-_31840438 | 0.07 |
ENST00000240587.4
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
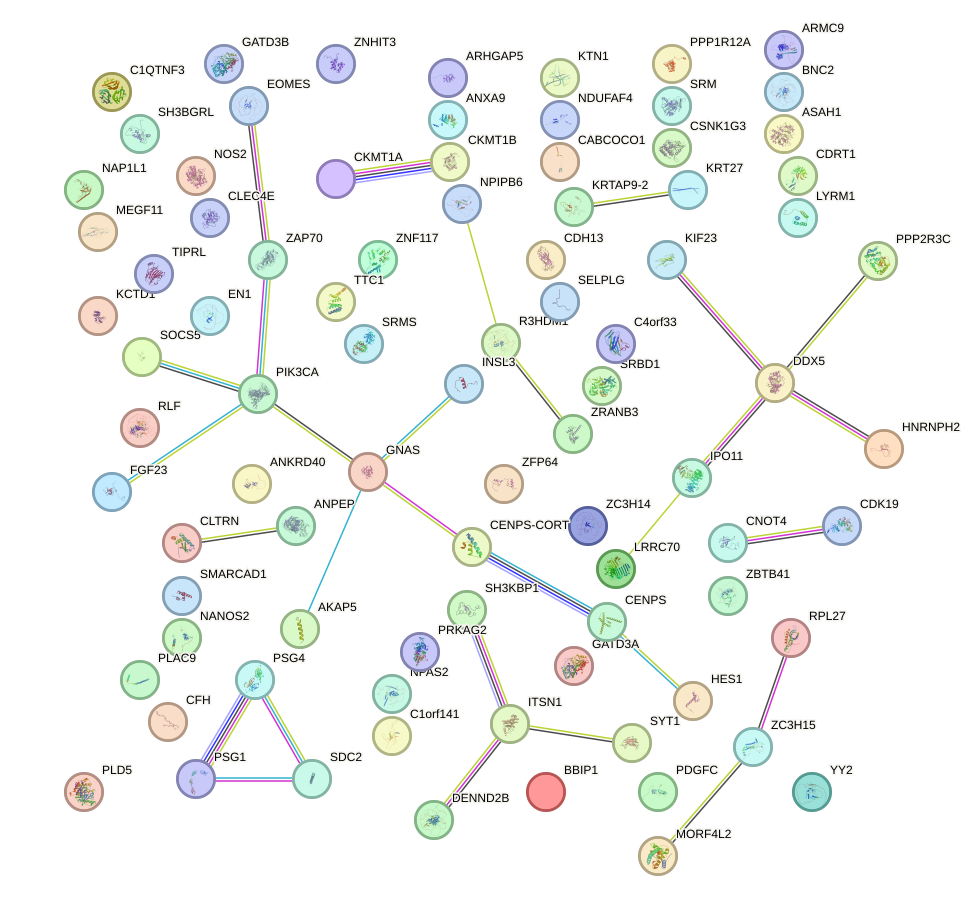
Network of associatons between targets according to the STRING database.
First level regulatory network of IKZF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.2 | GO:0060164 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) regulation of timing of neuron differentiation(GO:0060164) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.1 | 0.2 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.1 | 0.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.2 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.2 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.2 | GO:0042369 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) vitamin D catabolic process(GO:0042369) |
| 0.0 | 0.2 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.3 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.2 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.0 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.1 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.5 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.0 | 0.1 | GO:0072343 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.0 | 0.2 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0032618 | interleukin-15 production(GO:0032618) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.0 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.0 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.0 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.0 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.1 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) azole transporter activity(GO:0045118) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |