Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
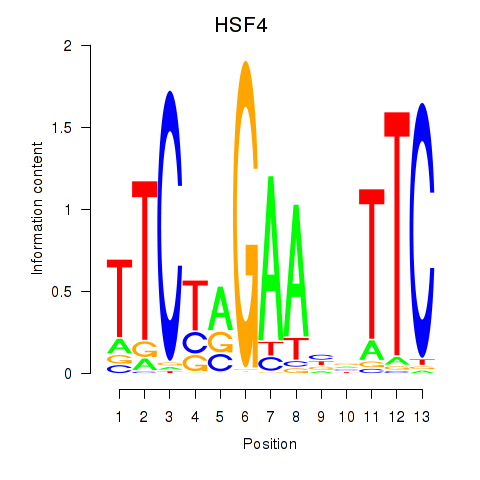
Results for HSF4
Z-value: 2.22
Transcription factors associated with HSF4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF4
|
ENSG00000102878.11 | heat shock transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF4 | hg19_v2_chr16_+_67198683_67198715 | -0.76 | 2.4e-01 | Click! |
Activity profile of HSF4 motif
Sorted Z-values of HSF4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_161494036 | 2.72 |
ENST00000309758.4
|
HSPA6
|
heat shock 70kDa protein 6 (HSP70B') |
| chrX_+_150866779 | 1.47 |
ENST00000370353.3
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr13_-_31736132 | 1.46 |
ENST00000429785.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr13_-_31736478 | 1.26 |
ENST00000445273.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chrX_+_108780062 | 1.07 |
ENST00000372106.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr7_+_157129738 | 1.02 |
ENST00000437030.1
|
DNAJB6
|
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chr13_-_31736027 | 1.01 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr7_-_1199781 | 1.00 |
ENST00000397083.1
ENST00000401903.1 ENST00000316495.3 |
ZFAND2A
|
zinc finger, AN1-type domain 2A |
| chr14_-_52535712 | 0.91 |
ENST00000216286.5
ENST00000541773.1 |
NID2
|
nidogen 2 (osteonidogen) |
| chr11_+_63953587 | 0.86 |
ENST00000305218.4
ENST00000538945.1 |
STIP1
|
stress-induced-phosphoprotein 1 |
| chr18_+_57567180 | 0.85 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr1_+_154947126 | 0.84 |
ENST00000368439.1
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr11_-_122932730 | 0.80 |
ENST00000532182.1
ENST00000524590.1 ENST00000528292.1 ENST00000533540.1 ENST00000525463.1 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr11_+_193065 | 0.80 |
ENST00000342878.2
|
SCGB1C1
|
secretoglobin, family 1C, member 1 |
| chr11_+_65266507 | 0.79 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr21_+_44394742 | 0.79 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr6_-_41168920 | 0.75 |
ENST00000483722.1
|
TREML2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr6_+_44215603 | 0.75 |
ENST00000371554.1
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr7_+_56119323 | 0.74 |
ENST00000275603.4
ENST00000335503.3 ENST00000540286.1 |
CCT6A
|
chaperonin containing TCP1, subunit 6A (zeta 1) |
| chr20_+_55108302 | 0.72 |
ENST00000371325.1
|
FAM209B
|
family with sequence similarity 209, member B |
| chr11_-_122933043 | 0.69 |
ENST00000534624.1
ENST00000453788.2 ENST00000527387.1 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr2_+_228678550 | 0.69 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr12_-_110939870 | 0.68 |
ENST00000447578.2
ENST00000546588.1 ENST00000360579.7 ENST00000549970.1 ENST00000549578.1 |
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr19_-_14628645 | 0.67 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr12_-_110883346 | 0.67 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr1_+_212782012 | 0.66 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
| chr8_+_62200509 | 0.64 |
ENST00000519846.1
ENST00000518592.1 ENST00000325897.4 |
CLVS1
|
clavesin 1 |
| chr1_+_154947148 | 0.62 |
ENST00000368436.1
ENST00000308987.5 |
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr11_+_66512303 | 0.61 |
ENST00000532565.2
|
C11orf80
|
chromosome 11 open reading frame 80 |
| chr1_-_6453426 | 0.59 |
ENST00000545482.1
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr12_+_104324112 | 0.58 |
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chrX_-_55515635 | 0.56 |
ENST00000500968.3
|
USP51
|
ubiquitin specific peptidase 51 |
| chr17_+_7258442 | 0.55 |
ENST00000389982.4
ENST00000576060.1 ENST00000330767.4 |
TMEM95
|
transmembrane protein 95 |
| chr19_+_3224700 | 0.54 |
ENST00000292672.2
ENST00000541430.2 |
CELF5
|
CUGBP, Elav-like family member 5 |
| chr14_+_63671105 | 0.52 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr18_-_6929829 | 0.51 |
ENST00000583840.1
ENST00000581571.1 ENST00000578497.1 ENST00000579012.1 |
LINC00668
|
long intergenic non-protein coding RNA 668 |
| chr19_+_45971246 | 0.51 |
ENST00000585836.1
ENST00000417353.2 ENST00000353609.3 ENST00000591858.1 ENST00000443841.2 ENST00000590335.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr1_+_92683467 | 0.51 |
ENST00000370375.3
ENST00000370373.2 |
C1orf146
|
chromosome 1 open reading frame 146 |
| chr17_+_74723031 | 0.51 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr9_+_74764340 | 0.49 |
ENST00000376986.1
ENST00000358399.3 |
GDA
|
guanine deaminase |
| chr15_+_79165222 | 0.49 |
ENST00000559930.1
|
MORF4L1
|
mortality factor 4 like 1 |
| chr12_-_122658746 | 0.49 |
ENST00000377035.1
|
IL31
|
interleukin 31 |
| chr11_-_134123142 | 0.49 |
ENST00000392595.2
ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1
|
thymocyte nuclear protein 1 |
| chr17_-_8286484 | 0.48 |
ENST00000582556.1
ENST00000584164.1 ENST00000293842.5 ENST00000584343.1 ENST00000578812.1 ENST00000583011.1 |
RPL26
|
ribosomal protein L26 |
| chr19_-_6720686 | 0.48 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr3_+_101443476 | 0.48 |
ENST00000327230.4
ENST00000494050.1 |
CEP97
|
centrosomal protein 97kDa |
| chr11_-_89956461 | 0.48 |
ENST00000320585.6
|
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr21_-_30445886 | 0.47 |
ENST00000431234.1
ENST00000540844.1 ENST00000286788.4 |
CCT8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr19_-_49567124 | 0.47 |
ENST00000301411.3
|
NTF4
|
neurotrophin 4 |
| chr2_+_201171064 | 0.46 |
ENST00000451764.2
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr2_+_201170999 | 0.46 |
ENST00000439395.1
ENST00000444012.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr18_+_11751466 | 0.46 |
ENST00000535121.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr7_+_26240776 | 0.46 |
ENST00000337620.4
|
CBX3
|
chromobox homolog 3 |
| chr12_+_2904102 | 0.46 |
ENST00000001008.4
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chrX_+_108779870 | 0.45 |
ENST00000372107.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr2_-_69180083 | 0.45 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chrX_-_128782722 | 0.45 |
ENST00000427399.1
|
APLN
|
apelin |
| chr17_+_34087888 | 0.45 |
ENST00000586491.1
ENST00000588628.1 ENST00000285023.4 |
C17orf50
|
chromosome 17 open reading frame 50 |
| chr13_+_31191920 | 0.45 |
ENST00000255304.4
|
USPL1
|
ubiquitin specific peptidase like 1 |
| chr19_-_14628234 | 0.45 |
ENST00000595139.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr5_+_172571445 | 0.44 |
ENST00000231668.9
ENST00000351486.5 ENST00000352523.6 ENST00000393770.4 |
BNIP1
|
BCL2/adenovirus E1B 19kDa interacting protein 1 |
| chr2_-_69664586 | 0.44 |
ENST00000303698.3
ENST00000394305.1 ENST00000410022.2 |
NFU1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr6_-_106773291 | 0.44 |
ENST00000343245.3
|
ATG5
|
autophagy related 5 |
| chr13_+_95254085 | 0.43 |
ENST00000376958.4
|
GPR180
|
G protein-coupled receptor 180 |
| chr3_-_53080644 | 0.43 |
ENST00000497586.1
|
SFMBT1
|
Scm-like with four mbt domains 1 |
| chr12_-_88423164 | 0.42 |
ENST00000298699.2
ENST00000550553.1 |
C12orf50
|
chromosome 12 open reading frame 50 |
| chr5_+_10250328 | 0.42 |
ENST00000515390.1
|
CCT5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr6_+_160693591 | 0.42 |
ENST00000419196.1
|
RP1-276N6.2
|
RP1-276N6.2 |
| chr11_+_66512089 | 0.42 |
ENST00000524551.1
ENST00000525908.1 ENST00000360962.4 ENST00000346672.4 ENST00000527634.1 ENST00000540737.1 |
C11orf80
|
chromosome 11 open reading frame 80 |
| chr18_-_71815051 | 0.42 |
ENST00000582526.1
ENST00000419743.2 |
FBXO15
|
F-box protein 15 |
| chr7_-_100493744 | 0.41 |
ENST00000428317.1
ENST00000441605.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr22_-_41252962 | 0.41 |
ENST00000216218.3
|
ST13
|
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) |
| chr1_-_156307992 | 0.41 |
ENST00000415548.1
|
CCT3
|
chaperonin containing TCP1, subunit 3 (gamma) |
| chr11_-_32816156 | 0.41 |
ENST00000531481.1
ENST00000335185.5 |
CCDC73
|
coiled-coil domain containing 73 |
| chr11_+_65769946 | 0.40 |
ENST00000533166.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr8_-_81083618 | 0.40 |
ENST00000520795.1
|
TPD52
|
tumor protein D52 |
| chr17_+_33474826 | 0.40 |
ENST00000268876.5
ENST00000433649.1 ENST00000378449.1 |
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr9_+_33025209 | 0.39 |
ENST00000330899.4
ENST00000544625.1 |
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr3_+_186501979 | 0.39 |
ENST00000498746.1
|
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr13_-_31191642 | 0.39 |
ENST00000405805.1
|
HMGB1
|
high mobility group box 1 |
| chr19_-_49565254 | 0.39 |
ENST00000593537.1
|
NTF4
|
neurotrophin 4 |
| chr12_+_70132632 | 0.38 |
ENST00000378815.6
ENST00000483530.2 ENST00000325555.9 |
RAB3IP
|
RAB3A interacting protein |
| chr7_-_56119238 | 0.38 |
ENST00000275605.3
ENST00000395471.3 |
PSPH
|
phosphoserine phosphatase |
| chrX_+_123095155 | 0.38 |
ENST00000371160.1
ENST00000435103.1 |
STAG2
|
stromal antigen 2 |
| chr22_+_32871224 | 0.38 |
ENST00000452138.1
ENST00000382058.3 ENST00000397426.1 |
FBXO7
|
F-box protein 7 |
| chr2_-_198364581 | 0.38 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr1_-_173638976 | 0.37 |
ENST00000333279.2
|
ANKRD45
|
ankyrin repeat domain 45 |
| chr16_-_18468926 | 0.37 |
ENST00000545114.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chrX_-_102942961 | 0.37 |
ENST00000434230.1
ENST00000418819.1 ENST00000360458.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr9_+_74764278 | 0.37 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr14_-_102553371 | 0.36 |
ENST00000553585.1
ENST00000216281.8 |
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr8_+_143781883 | 0.36 |
ENST00000522591.1
|
LY6K
|
lymphocyte antigen 6 complex, locus K |
| chr3_+_130613001 | 0.36 |
ENST00000504948.1
ENST00000513801.1 ENST00000505072.1 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr22_+_17082732 | 0.36 |
ENST00000558085.2
ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1
|
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr11_-_82997420 | 0.36 |
ENST00000455220.2
ENST00000529689.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr2_+_201170770 | 0.36 |
ENST00000409988.3
ENST00000409385.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr8_+_37594130 | 0.36 |
ENST00000518526.1
ENST00000523887.1 ENST00000276461.5 |
ERLIN2
|
ER lipid raft associated 2 |
| chr11_-_89956227 | 0.36 |
ENST00000457199.2
ENST00000530765.1 |
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr2_+_198365095 | 0.35 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr8_+_140943416 | 0.35 |
ENST00000507535.3
|
C8orf17
|
chromosome 8 open reading frame 17 |
| chr8_-_81083731 | 0.34 |
ENST00000379096.5
|
TPD52
|
tumor protein D52 |
| chrX_-_154563889 | 0.34 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr7_-_3083573 | 0.34 |
ENST00000396946.4
|
CARD11
|
caspase recruitment domain family, member 11 |
| chr3_+_130613226 | 0.34 |
ENST00000509662.1
ENST00000328560.8 ENST00000428331.2 ENST00000359644.3 ENST00000422190.2 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chrX_+_123094369 | 0.34 |
ENST00000455404.1
ENST00000218089.9 |
STAG2
|
stromal antigen 2 |
| chr14_+_101293687 | 0.33 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr21_+_35445811 | 0.33 |
ENST00000399312.2
|
MRPS6
|
mitochondrial ribosomal protein S6 |
| chr7_+_157129660 | 0.33 |
ENST00000429029.2
ENST00000262177.4 ENST00000417758.1 ENST00000452797.2 ENST00000443280.1 |
DNAJB6
|
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chrX_-_102943022 | 0.33 |
ENST00000433176.2
|
MORF4L2
|
mortality factor 4 like 2 |
| chr9_-_88714421 | 0.32 |
ENST00000388712.3
|
GOLM1
|
golgi membrane protein 1 |
| chr20_-_43589109 | 0.32 |
ENST00000372813.3
|
TOMM34
|
translocase of outer mitochondrial membrane 34 |
| chr17_+_33474860 | 0.32 |
ENST00000394570.2
|
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr1_-_27709793 | 0.32 |
ENST00000374027.3
ENST00000374025.3 |
CD164L2
|
CD164 sialomucin-like 2 |
| chr6_-_138833630 | 0.31 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr5_+_133706865 | 0.31 |
ENST00000265339.2
|
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr22_+_31460091 | 0.31 |
ENST00000432777.1
ENST00000422839.1 |
SMTN
|
smoothelin |
| chr1_-_26232522 | 0.31 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr2_-_62115659 | 0.31 |
ENST00000544185.1
|
CCT4
|
chaperonin containing TCP1, subunit 4 (delta) |
| chr2_+_201171242 | 0.31 |
ENST00000360760.5
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr11_-_111781554 | 0.30 |
ENST00000526167.1
ENST00000528961.1 |
CRYAB
|
crystallin, alpha B |
| chr4_-_17513604 | 0.30 |
ENST00000505710.1
|
QDPR
|
quinoid dihydropteridine reductase |
| chr16_-_68034470 | 0.29 |
ENST00000412757.2
|
DPEP2
|
dipeptidase 2 |
| chr5_+_118690466 | 0.29 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr6_+_36165133 | 0.29 |
ENST00000446974.1
ENST00000454960.1 |
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chr1_-_149459549 | 0.29 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr17_-_74722536 | 0.29 |
ENST00000585429.1
|
JMJD6
|
jumonji domain containing 6 |
| chr3_+_136676851 | 0.28 |
ENST00000309741.5
|
IL20RB
|
interleukin 20 receptor beta |
| chr21_+_35445827 | 0.27 |
ENST00000608209.1
ENST00000381151.3 |
SLC5A3
SLC5A3
|
sodium/myo-inositol cotransporter solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
| chr17_-_62308087 | 0.26 |
ENST00000583097.1
|
TEX2
|
testis expressed 2 |
| chr1_+_156308403 | 0.25 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr9_-_88715044 | 0.25 |
ENST00000388711.3
ENST00000466178.1 |
GOLM1
|
golgi membrane protein 1 |
| chr11_-_14913765 | 0.25 |
ENST00000334636.5
|
CYP2R1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr22_+_41253080 | 0.25 |
ENST00000541156.1
ENST00000414396.1 ENST00000357137.4 |
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chr5_+_159895275 | 0.25 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr11_+_112046190 | 0.25 |
ENST00000357685.5
ENST00000393032.2 ENST00000361053.4 |
BCO2
|
beta-carotene oxygenase 2 |
| chr8_+_37594185 | 0.24 |
ENST00000518586.1
ENST00000335171.6 ENST00000521644.1 |
ERLIN2
|
ER lipid raft associated 2 |
| chr2_+_201171268 | 0.24 |
ENST00000423749.1
ENST00000428692.1 ENST00000457757.1 ENST00000453663.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr17_-_46716647 | 0.24 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr1_+_67773044 | 0.24 |
ENST00000262345.1
ENST00000371000.1 |
IL12RB2
|
interleukin 12 receptor, beta 2 |
| chr15_+_79165112 | 0.24 |
ENST00000426013.2
|
MORF4L1
|
mortality factor 4 like 1 |
| chr11_-_47870019 | 0.23 |
ENST00000378460.2
|
NUP160
|
nucleoporin 160kDa |
| chr11_-_62457371 | 0.23 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr3_-_183735731 | 0.23 |
ENST00000334444.6
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chrX_+_123094672 | 0.23 |
ENST00000354548.5
ENST00000458700.1 |
STAG2
|
stromal antigen 2 |
| chr22_+_50986462 | 0.22 |
ENST00000395676.2
|
KLHDC7B
|
kelch domain containing 7B |
| chr3_-_53080672 | 0.22 |
ENST00000483069.1
|
SFMBT1
|
Scm-like with four mbt domains 1 |
| chrX_-_152939252 | 0.21 |
ENST00000340888.3
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr3_+_130612803 | 0.21 |
ENST00000510168.1
ENST00000508532.1 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr2_+_203130439 | 0.21 |
ENST00000264279.5
|
NOP58
|
NOP58 ribonucleoprotein |
| chr15_+_79165151 | 0.21 |
ENST00000331268.5
|
MORF4L1
|
mortality factor 4 like 1 |
| chrX_-_39923656 | 0.21 |
ENST00000413905.1
|
BCOR
|
BCL6 corepressor |
| chr18_-_19284724 | 0.21 |
ENST00000580981.1
ENST00000289119.2 |
ABHD3
|
abhydrolase domain containing 3 |
| chr19_+_1354930 | 0.21 |
ENST00000591337.1
|
MUM1
|
melanoma associated antigen (mutated) 1 |
| chr21_-_40720974 | 0.21 |
ENST00000380748.1
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr21_-_40720995 | 0.20 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chrX_-_152939133 | 0.20 |
ENST00000370150.1
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr8_-_37594944 | 0.20 |
ENST00000330539.1
|
RP11-863K10.7
|
Uncharacterized protein |
| chr3_-_185655795 | 0.20 |
ENST00000342294.4
ENST00000382191.4 ENST00000453386.2 |
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr17_-_61850894 | 0.20 |
ENST00000403162.3
ENST00000582252.1 ENST00000225726.5 |
CCDC47
|
coiled-coil domain containing 47 |
| chr11_+_107799118 | 0.19 |
ENST00000320578.2
|
RAB39A
|
RAB39A, member RAS oncogene family |
| chr11_-_111781454 | 0.19 |
ENST00000533280.1
|
CRYAB
|
crystallin, alpha B |
| chr1_+_109265033 | 0.19 |
ENST00000445274.1
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr12_-_48500085 | 0.19 |
ENST00000549518.1
|
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr3_+_136676707 | 0.19 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr1_-_248005254 | 0.19 |
ENST00000355784.2
|
OR11L1
|
olfactory receptor, family 11, subfamily L, member 1 |
| chr17_-_74722672 | 0.19 |
ENST00000397625.4
ENST00000445478.2 |
JMJD6
|
jumonji domain containing 6 |
| chrX_-_106243451 | 0.18 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr10_-_74856608 | 0.18 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr19_+_2269485 | 0.18 |
ENST00000582888.4
ENST00000602676.2 ENST00000322297.4 ENST00000583542.4 |
OAZ1
|
ornithine decarboxylase antizyme 1 |
| chr19_+_41509851 | 0.18 |
ENST00000593831.1
ENST00000330446.5 |
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr5_+_133707252 | 0.18 |
ENST00000506787.1
ENST00000507277.1 |
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr2_+_201170703 | 0.17 |
ENST00000358677.5
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr5_+_56469843 | 0.17 |
ENST00000514387.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr18_+_11751493 | 0.17 |
ENST00000269162.5
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr2_+_201171577 | 0.17 |
ENST00000409397.2
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr11_-_111782696 | 0.17 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr5_+_56469939 | 0.17 |
ENST00000506184.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr16_+_14844670 | 0.16 |
ENST00000553201.1
|
NPIPA2
|
nuclear pore complex interacting protein family, member A2 |
| chr12_+_70132458 | 0.16 |
ENST00000247833.7
|
RAB3IP
|
RAB3A interacting protein |
| chr4_-_22517620 | 0.16 |
ENST00000502482.1
ENST00000334304.5 |
GPR125
|
G protein-coupled receptor 125 |
| chr4_-_6383594 | 0.16 |
ENST00000335585.5
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr4_-_159644507 | 0.16 |
ENST00000307720.3
|
PPID
|
peptidylprolyl isomerase D |
| chr19_-_4717835 | 0.16 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr2_-_68479614 | 0.16 |
ENST00000234310.3
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chrX_-_106243294 | 0.16 |
ENST00000255495.7
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr19_+_11650709 | 0.16 |
ENST00000586059.1
|
CNN1
|
calponin 1, basic, smooth muscle |
| chr12_-_123011536 | 0.16 |
ENST00000331738.7
ENST00000354654.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr19_+_6135646 | 0.16 |
ENST00000588304.1
ENST00000588485.1 ENST00000588722.1 ENST00000591403.1 ENST00000586696.1 ENST00000589401.1 ENST00000252669.5 |
ACSBG2
|
acyl-CoA synthetase bubblegum family member 2 |
| chrX_-_73072534 | 0.16 |
ENST00000429829.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr15_+_45028753 | 0.15 |
ENST00000338264.4
|
TRIM69
|
tripartite motif containing 69 |
| chrX_-_152989798 | 0.15 |
ENST00000441714.1
ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31
|
B-cell receptor-associated protein 31 |
| chr20_+_4666882 | 0.15 |
ENST00000379440.4
ENST00000430350.2 |
PRNP
|
prion protein |
| chr20_-_23731569 | 0.15 |
ENST00000304749.2
|
CST1
|
cystatin SN |
| chr14_-_68141535 | 0.15 |
ENST00000554659.1
|
VTI1B
|
vesicle transport through interaction with t-SNAREs 1B |
| chr12_-_123011476 | 0.15 |
ENST00000528279.1
ENST00000344591.4 ENST00000526560.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr11_+_63953691 | 0.14 |
ENST00000543847.1
|
STIP1
|
stress-induced-phosphoprotein 1 |
| chr11_-_82997371 | 0.14 |
ENST00000525503.1
|
CCDC90B
|
coiled-coil domain containing 90B |
| chr1_-_45987526 | 0.13 |
ENST00000372079.1
ENST00000262746.1 ENST00000447184.1 ENST00000319248.8 |
PRDX1
|
peroxiredoxin 1 |
| chr1_-_245026388 | 0.13 |
ENST00000440865.1
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr9_-_86323118 | 0.13 |
ENST00000376395.4
|
UBQLN1
|
ubiquilin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.7 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.4 | 2.7 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.3 | 1.5 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.3 | 0.9 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.3 | 0.9 | GO:0008052 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.2 | 0.7 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.2 | 0.7 | GO:0045362 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 0.7 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.2 | 0.8 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.2 | 0.6 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.2 | 0.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.2 | 0.7 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 | 0.5 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.2 | 0.5 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.2 | 2.5 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 0.4 | GO:0051466 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.1 | 1.3 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.1 | 0.4 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.9 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.5 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.1 | 0.4 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.1 | 0.4 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.1 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 1.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.6 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.4 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.4 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.5 | GO:1902231 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.5 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.9 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.4 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.1 | 0.2 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.1 | 0.8 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.1 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 1.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.5 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.2 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.8 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.3 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.4 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.3 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.5 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.5 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.2 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.9 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.5 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.2 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.4 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.4 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.2 | 1.5 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.2 | 4.7 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.7 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 2.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.4 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.4 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 2.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 1.5 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.3 | 3.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 0.7 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.2 | 1.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 0.7 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.2 | 1.9 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.9 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.4 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.1 | 0.5 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.4 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.4 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.3 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.1 | 0.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.5 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.4 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.7 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.2 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.4 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.2 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 7.8 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.2 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0004028 | retinal dehydrogenase activity(GO:0001758) 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.0 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.9 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.5 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |