Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
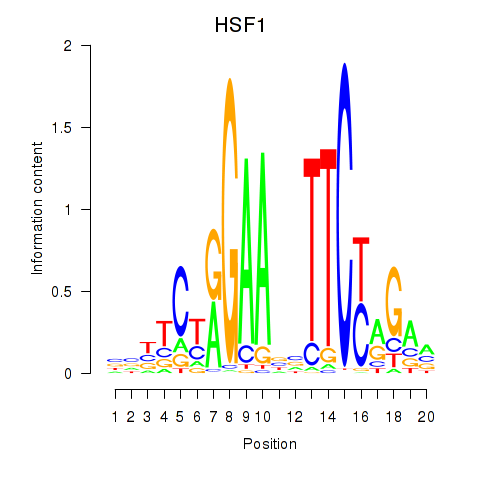
Results for HSF1
Z-value: 0.31
Transcription factors associated with HSF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF1
|
ENSG00000185122.6 | heat shock transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF1 | hg19_v2_chr8_+_145515263_145515299 | -0.89 | 1.1e-01 | Click! |
Activity profile of HSF1 motif
Sorted Z-values of HSF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_15001430 | 0.25 |
ENST00000407572.1
|
MEIG1
|
meiosis/spermiogenesis associated 1 |
| chr20_-_44455976 | 0.18 |
ENST00000372555.3
|
TNNC2
|
troponin C type 2 (fast) |
| chr1_+_161494036 | 0.17 |
ENST00000309758.4
|
HSPA6
|
heat shock 70kDa protein 6 (HSP70B') |
| chrX_+_150866779 | 0.16 |
ENST00000370353.3
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr7_+_157129738 | 0.16 |
ENST00000437030.1
|
DNAJB6
|
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chr17_+_7258442 | 0.12 |
ENST00000389982.4
ENST00000576060.1 ENST00000330767.4 |
TMEM95
|
transmembrane protein 95 |
| chr18_+_57567180 | 0.12 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr2_-_27851745 | 0.11 |
ENST00000394775.3
ENST00000522876.1 |
CCDC121
|
coiled-coil domain containing 121 |
| chr20_-_57617831 | 0.11 |
ENST00000371033.5
ENST00000355937.4 |
SLMO2
|
slowmo homolog 2 (Drosophila) |
| chr3_+_46742823 | 0.11 |
ENST00000326431.3
|
TMIE
|
transmembrane inner ear |
| chr1_-_42630389 | 0.11 |
ENST00000357001.2
|
GUCA2A
|
guanylate cyclase activator 2A (guanylin) |
| chr1_-_109584716 | 0.10 |
ENST00000531337.1
ENST00000529074.1 ENST00000369965.4 |
WDR47
|
WD repeat domain 47 |
| chrX_+_108780062 | 0.10 |
ENST00000372106.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr10_+_71444655 | 0.10 |
ENST00000434931.2
|
RP11-242G20.1
|
Uncharacterized protein |
| chr19_-_10311868 | 0.10 |
ENST00000588118.1
ENST00000586800.1 |
DNMT1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr1_+_92683467 | 0.10 |
ENST00000370375.3
ENST00000370373.2 |
C1orf146
|
chromosome 1 open reading frame 146 |
| chr11_-_134123142 | 0.10 |
ENST00000392595.2
ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1
|
thymocyte nuclear protein 1 |
| chr2_-_27851843 | 0.10 |
ENST00000324364.3
|
CCDC121
|
coiled-coil domain containing 121 |
| chr8_-_56986768 | 0.10 |
ENST00000523936.1
|
RPS20
|
ribosomal protein S20 |
| chr17_-_58499766 | 0.10 |
ENST00000588898.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr9_-_34523027 | 0.10 |
ENST00000399775.2
|
ENHO
|
energy homeostasis associated |
| chr6_-_31782813 | 0.09 |
ENST00000375654.4
|
HSPA1L
|
heat shock 70kDa protein 1-like |
| chr4_+_57396766 | 0.09 |
ENST00000512175.2
|
THEGL
|
theg spermatid protein-like |
| chr14_+_100532771 | 0.09 |
ENST00000557153.1
|
EVL
|
Enah/Vasp-like |
| chr2_-_151395525 | 0.09 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr11_-_89956227 | 0.09 |
ENST00000457199.2
ENST00000530765.1 |
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr3_-_107941230 | 0.08 |
ENST00000264538.3
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr17_+_41005283 | 0.08 |
ENST00000592999.1
|
AOC3
|
amine oxidase, copper containing 3 |
| chr1_+_212782012 | 0.08 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
| chr3_+_119421849 | 0.08 |
ENST00000273390.5
ENST00000463700.1 |
MAATS1
|
MYCBP-associated, testis expressed 1 |
| chr1_-_27682962 | 0.08 |
ENST00000486046.1
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr8_-_56987061 | 0.08 |
ENST00000009589.3
ENST00000524349.1 ENST00000519606.1 ENST00000519807.1 ENST00000521262.1 ENST00000520627.1 |
RPS20
|
ribosomal protein S20 |
| chr20_+_43803517 | 0.08 |
ENST00000243924.3
|
PI3
|
peptidase inhibitor 3, skin-derived |
| chr6_+_32937083 | 0.08 |
ENST00000456339.1
|
BRD2
|
bromodomain containing 2 |
| chr6_+_33551515 | 0.08 |
ENST00000374458.1
|
GGNBP1
|
gametogenetin binding protein 1 (pseudogene) |
| chr16_+_222846 | 0.08 |
ENST00000251595.6
ENST00000397806.1 |
HBA2
|
hemoglobin, alpha 2 |
| chr11_-_89956461 | 0.08 |
ENST00000320585.6
|
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr8_-_144413981 | 0.08 |
ENST00000522041.1
|
TOP1MT
|
topoisomerase (DNA) I, mitochondrial |
| chr9_-_98079154 | 0.08 |
ENST00000433829.1
|
FANCC
|
Fanconi anemia, complementation group C |
| chr16_-_75529273 | 0.08 |
ENST00000390664.2
|
CHST6
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6 |
| chr9_+_127615733 | 0.08 |
ENST00000373574.1
|
WDR38
|
WD repeat domain 38 |
| chr1_+_212738676 | 0.07 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr5_+_32531893 | 0.07 |
ENST00000512913.1
|
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr15_-_89764929 | 0.07 |
ENST00000268125.5
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr12_-_123011476 | 0.07 |
ENST00000528279.1
ENST00000344591.4 ENST00000526560.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr10_-_101673782 | 0.07 |
ENST00000422692.1
|
DNMBP
|
dynamin binding protein |
| chr6_+_10694900 | 0.07 |
ENST00000379568.3
|
PAK1IP1
|
PAK1 interacting protein 1 |
| chr1_+_111415757 | 0.07 |
ENST00000429072.2
ENST00000271324.5 |
CD53
|
CD53 molecule |
| chrX_+_108779870 | 0.07 |
ENST00000372107.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr4_+_166128836 | 0.07 |
ENST00000511305.1
|
KLHL2
|
kelch-like family member 2 |
| chr9_-_113018746 | 0.07 |
ENST00000374515.5
|
TXN
|
thioredoxin |
| chr5_-_76788134 | 0.07 |
ENST00000507029.1
|
WDR41
|
WD repeat domain 41 |
| chr12_-_76742183 | 0.07 |
ENST00000393262.3
|
BBS10
|
Bardet-Biedl syndrome 10 |
| chr6_-_160210604 | 0.07 |
ENST00000420894.2
ENST00000539756.1 ENST00000544255.1 |
TCP1
|
t-complex 1 |
| chr13_+_31191920 | 0.07 |
ENST00000255304.4
|
USPL1
|
ubiquitin specific peptidase like 1 |
| chr2_+_198365095 | 0.07 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr2_+_27851863 | 0.07 |
ENST00000264718.3
ENST00000610189.1 |
GPN1
|
GPN-loop GTPase 1 |
| chr4_+_76481258 | 0.07 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr11_+_60145997 | 0.07 |
ENST00000530614.1
ENST00000530027.1 ENST00000530234.2 ENST00000528215.1 ENST00000531787.1 |
MS4A7
MS4A14
|
membrane-spanning 4-domains, subfamily A, member 7 membrane-spanning 4-domains, subfamily A, member 14 |
| chr15_+_79165222 | 0.07 |
ENST00000559930.1
|
MORF4L1
|
mortality factor 4 like 1 |
| chrX_-_108976521 | 0.07 |
ENST00000469796.2
ENST00000502391.1 ENST00000508092.1 ENST00000340800.2 ENST00000348502.6 |
ACSL4
|
acyl-CoA synthetase long-chain family member 4 |
| chr2_+_237476419 | 0.07 |
ENST00000447924.1
|
ACKR3
|
atypical chemokine receptor 3 |
| chrX_-_108976410 | 0.07 |
ENST00000504980.1
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4 |
| chr18_-_51751132 | 0.07 |
ENST00000256429.3
|
MBD2
|
methyl-CpG binding domain protein 2 |
| chr2_-_198364581 | 0.07 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr15_+_78857849 | 0.06 |
ENST00000299565.5
|
CHRNA5
|
cholinergic receptor, nicotinic, alpha 5 (neuronal) |
| chr5_+_158737824 | 0.06 |
ENST00000521472.1
|
AC008697.1
|
AC008697.1 |
| chr19_-_14628645 | 0.06 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr8_-_134511587 | 0.06 |
ENST00000523855.1
ENST00000523854.1 |
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr4_+_119200215 | 0.06 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr11_-_32816156 | 0.06 |
ENST00000531481.1
ENST00000335185.5 |
CCDC73
|
coiled-coil domain containing 73 |
| chr5_+_178450753 | 0.06 |
ENST00000444149.2
ENST00000519896.1 ENST00000522442.1 |
ZNF879
|
zinc finger protein 879 |
| chr8_+_56685701 | 0.06 |
ENST00000260129.5
|
TGS1
|
trimethylguanosine synthase 1 |
| chr9_+_72658490 | 0.06 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr3_-_117716418 | 0.06 |
ENST00000484092.1
|
RP11-384F7.2
|
RP11-384F7.2 |
| chr2_-_201729284 | 0.06 |
ENST00000434813.2
|
CLK1
|
CDC-like kinase 1 |
| chr2_+_103089756 | 0.06 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chrX_+_37208521 | 0.06 |
ENST00000378628.4
|
PRRG1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr11_-_102323489 | 0.06 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr1_-_143913143 | 0.06 |
ENST00000400889.1
|
FAM72D
|
family with sequence similarity 72, member D |
| chr6_-_105627735 | 0.06 |
ENST00000254765.3
|
POPDC3
|
popeye domain containing 3 |
| chr11_+_33563821 | 0.06 |
ENST00000321505.4
ENST00000265654.5 ENST00000389726.3 |
KIAA1549L
|
KIAA1549-like |
| chr7_-_151433342 | 0.06 |
ENST00000433631.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr11_-_64163297 | 0.06 |
ENST00000457725.1
|
AP003774.6
|
AP003774.6 |
| chr8_-_81083341 | 0.06 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chr16_+_85942594 | 0.06 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8 |
| chr8_+_124194752 | 0.06 |
ENST00000318462.6
|
FAM83A
|
family with sequence similarity 83, member A |
| chr11_+_64323098 | 0.06 |
ENST00000301891.4
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr6_-_46620522 | 0.06 |
ENST00000275016.2
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr11_-_125773085 | 0.06 |
ENST00000227474.3
ENST00000534158.1 ENST00000529801.1 |
PUS3
|
pseudouridylate synthase 3 |
| chr3_+_101443476 | 0.06 |
ENST00000327230.4
ENST00000494050.1 |
CEP97
|
centrosomal protein 97kDa |
| chr8_-_81083890 | 0.05 |
ENST00000518937.1
|
TPD52
|
tumor protein D52 |
| chr6_+_35265586 | 0.05 |
ENST00000542066.1
ENST00000316637.5 |
DEF6
|
differentially expressed in FDCP 6 homolog (mouse) |
| chr11_+_65851443 | 0.05 |
ENST00000533756.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr12_-_10826612 | 0.05 |
ENST00000535345.1
ENST00000542562.1 |
STYK1
|
serine/threonine/tyrosine kinase 1 |
| chr19_-_14217672 | 0.05 |
ENST00000587372.1
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha |
| chrX_-_128782722 | 0.05 |
ENST00000427399.1
|
APLN
|
apelin |
| chr18_+_44526786 | 0.05 |
ENST00000245121.5
ENST00000356157.7 |
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr3_+_148847371 | 0.05 |
ENST00000296051.2
ENST00000460120.1 |
HPS3
|
Hermansky-Pudlak syndrome 3 |
| chr2_-_202316260 | 0.05 |
ENST00000332624.3
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr17_+_53828432 | 0.05 |
ENST00000573500.1
|
PCTP
|
phosphatidylcholine transfer protein |
| chr15_+_78857870 | 0.05 |
ENST00000559554.1
|
CHRNA5
|
cholinergic receptor, nicotinic, alpha 5 (neuronal) |
| chr18_-_52626622 | 0.05 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr18_+_44526744 | 0.05 |
ENST00000585469.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr19_-_12444306 | 0.05 |
ENST00000594577.1
ENST00000601858.1 |
ZNF563
|
zinc finger protein 563 |
| chr3_+_56591184 | 0.05 |
ENST00000422222.1
ENST00000394672.3 ENST00000326595.7 |
CCDC66
|
coiled-coil domain containing 66 |
| chrX_+_37208540 | 0.05 |
ENST00000466533.1
ENST00000542554.1 ENST00000543642.1 ENST00000484460.1 ENST00000449135.2 ENST00000463135.1 ENST00000465127.1 |
PRRG1
TM4SF2
|
proline rich Gla (G-carboxyglutamic acid) 1 Uncharacterized protein; cDNA FLJ59144, highly similar to Tetraspanin-7 |
| chr10_-_33281363 | 0.05 |
ENST00000534049.1
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
| chr1_+_120839005 | 0.05 |
ENST00000369390.3
ENST00000452190.1 |
FAM72B
|
family with sequence similarity 72, member B |
| chr6_-_100016527 | 0.05 |
ENST00000523985.1
ENST00000518714.1 ENST00000520371.1 |
CCNC
|
cyclin C |
| chr1_-_109584768 | 0.05 |
ENST00000357672.3
|
WDR47
|
WD repeat domain 47 |
| chr6_-_160210715 | 0.05 |
ENST00000392168.2
ENST00000321394.7 |
TCP1
|
t-complex 1 |
| chr9_+_127054254 | 0.05 |
ENST00000422297.1
|
NEK6
|
NIMA-related kinase 6 |
| chr3_+_130613226 | 0.05 |
ENST00000509662.1
ENST00000328560.8 ENST00000428331.2 ENST00000359644.3 ENST00000422190.2 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr6_+_168434678 | 0.05 |
ENST00000496008.1
|
KIF25
|
kinesin family member 25 |
| chr6_-_108582168 | 0.05 |
ENST00000426155.2
|
SNX3
|
sorting nexin 3 |
| chr3_-_142720267 | 0.05 |
ENST00000597953.1
|
RP11-91G21.1
|
RP11-91G21.1 |
| chr1_-_224517823 | 0.05 |
ENST00000469968.1
ENST00000436927.1 ENST00000469075.1 ENST00000488718.1 ENST00000482491.1 ENST00000340871.4 ENST00000492281.1 ENST00000361463.3 ENST00000391875.2 ENST00000461546.1 |
NVL
|
nuclear VCP-like |
| chr9_+_74764278 | 0.05 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr14_+_63671105 | 0.05 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr14_+_100789669 | 0.05 |
ENST00000361529.3
ENST00000557052.1 |
SLC25A47
|
solute carrier family 25, member 47 |
| chr18_-_32870148 | 0.05 |
ENST00000589178.1
ENST00000333206.5 ENST00000592278.1 ENST00000592211.1 ENST00000420878.3 ENST00000383091.2 ENST00000586922.2 |
ZSCAN30
RP11-158H5.7
|
zinc finger and SCAN domain containing 30 RP11-158H5.7 |
| chr10_+_103348031 | 0.05 |
ENST00000370151.4
ENST00000370147.1 ENST00000370148.2 |
DPCD
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chrX_+_100663243 | 0.05 |
ENST00000316594.5
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr14_+_31028348 | 0.05 |
ENST00000550944.1
ENST00000438909.2 ENST00000553504.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr7_+_26241310 | 0.05 |
ENST00000396386.2
|
CBX3
|
chromobox homolog 3 |
| chr8_+_96037255 | 0.05 |
ENST00000286687.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chrX_-_39923656 | 0.05 |
ENST00000413905.1
|
BCOR
|
BCL6 corepressor |
| chr1_-_45272951 | 0.05 |
ENST00000372200.1
|
TCTEX1D4
|
Tctex1 domain containing 4 |
| chr3_+_130613001 | 0.05 |
ENST00000504948.1
ENST00000513801.1 ENST00000505072.1 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr22_-_41252962 | 0.05 |
ENST00000216218.3
|
ST13
|
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) |
| chr14_-_50319482 | 0.05 |
ENST00000546046.1
ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF
|
nuclear export mediator factor |
| chr9_-_86432547 | 0.05 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr7_-_102715263 | 0.05 |
ENST00000379305.3
|
FBXL13
|
F-box and leucine-rich repeat protein 13 |
| chr1_+_19970202 | 0.05 |
ENST00000439664.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr2_-_218867711 | 0.05 |
ENST00000446903.1
|
TNS1
|
tensin 1 |
| chr4_-_147442817 | 0.05 |
ENST00000507030.1
|
SLC10A7
|
solute carrier family 10, member 7 |
| chr7_-_100493744 | 0.05 |
ENST00000428317.1
ENST00000441605.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr2_+_27851106 | 0.05 |
ENST00000515877.1
|
GPN1
|
GPN-loop GTPase 1 |
| chr6_-_106773291 | 0.05 |
ENST00000343245.3
|
ATG5
|
autophagy related 5 |
| chr19_-_14628234 | 0.05 |
ENST00000595139.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr19_+_11650709 | 0.05 |
ENST00000586059.1
|
CNN1
|
calponin 1, basic, smooth muscle |
| chr16_-_18468926 | 0.05 |
ENST00000545114.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr12_+_104324112 | 0.05 |
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr12_+_93772326 | 0.05 |
ENST00000550056.1
ENST00000549992.1 ENST00000548662.1 ENST00000547014.1 |
NUDT4
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr2_-_87248975 | 0.05 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr17_-_60005365 | 0.05 |
ENST00000444766.3
|
INTS2
|
integrator complex subunit 2 |
| chr7_+_157129660 | 0.05 |
ENST00000429029.2
ENST00000262177.4 ENST00000417758.1 ENST00000452797.2 ENST00000443280.1 |
DNAJB6
|
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chr16_-_53537105 | 0.05 |
ENST00000568596.1
ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP
|
AKT interacting protein |
| chr6_-_131277510 | 0.05 |
ENST00000525193.1
ENST00000527659.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chrX_-_152939133 | 0.04 |
ENST00000370150.1
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr9_-_88715044 | 0.04 |
ENST00000388711.3
ENST00000466178.1 |
GOLM1
|
golgi membrane protein 1 |
| chr21_+_33031935 | 0.04 |
ENST00000270142.6
ENST00000389995.4 |
SOD1
|
superoxide dismutase 1, soluble |
| chr22_+_29664241 | 0.04 |
ENST00000436425.1
ENST00000447973.1 |
EWSR1
|
EWS RNA-binding protein 1 |
| chr14_+_31028329 | 0.04 |
ENST00000206595.6
|
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr11_-_94706705 | 0.04 |
ENST00000279839.6
|
CWC15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr3_+_154797781 | 0.04 |
ENST00000382989.3
|
MME
|
membrane metallo-endopeptidase |
| chr2_+_202316392 | 0.04 |
ENST00000194530.3
ENST00000392249.2 |
STRADB
|
STE20-related kinase adaptor beta |
| chr4_+_106816644 | 0.04 |
ENST00000506666.1
ENST00000503451.1 |
NPNT
|
nephronectin |
| chr1_+_1260598 | 0.04 |
ENST00000488011.1
|
GLTPD1
|
glycolipid transfer protein domain containing 1 |
| chr4_+_190861941 | 0.04 |
ENST00000226798.4
|
FRG1
|
FSHD region gene 1 |
| chr2_+_135011731 | 0.04 |
ENST00000281923.2
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr8_-_56685859 | 0.04 |
ENST00000523423.1
ENST00000523073.1 ENST00000519784.1 ENST00000434581.2 ENST00000519780.1 ENST00000521229.1 ENST00000522576.1 ENST00000523180.1 ENST00000522090.1 |
TMEM68
|
transmembrane protein 68 |
| chr13_-_31191642 | 0.04 |
ENST00000405805.1
|
HMGB1
|
high mobility group box 1 |
| chr2_-_56150910 | 0.04 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr4_-_89619386 | 0.04 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr10_-_16859361 | 0.04 |
ENST00000377921.3
|
RSU1
|
Ras suppressor protein 1 |
| chr16_+_28996572 | 0.04 |
ENST00000360872.5
ENST00000566177.1 ENST00000354453.4 |
LAT
|
linker for activation of T cells |
| chr4_-_84406307 | 0.04 |
ENST00000321945.7
|
FAM175A
|
family with sequence similarity 175, member A |
| chr12_+_111051832 | 0.04 |
ENST00000550703.2
ENST00000551590.1 |
TCTN1
|
tectonic family member 1 |
| chr2_+_109403193 | 0.04 |
ENST00000412964.2
ENST00000295124.4 |
CCDC138
|
coiled-coil domain containing 138 |
| chr19_-_55660561 | 0.04 |
ENST00000587758.1
ENST00000356783.5 ENST00000291901.8 ENST00000588426.1 ENST00000588147.1 ENST00000536926.1 ENST00000588981.1 |
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr9_+_71736177 | 0.04 |
ENST00000606364.1
ENST00000453658.2 |
TJP2
|
tight junction protein 2 |
| chr2_-_191878874 | 0.04 |
ENST00000392322.3
ENST00000392323.2 ENST00000424722.1 ENST00000361099.3 |
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr8_-_17104356 | 0.04 |
ENST00000361272.4
ENST00000523917.1 |
CNOT7
|
CCR4-NOT transcription complex, subunit 7 |
| chr2_-_234763105 | 0.04 |
ENST00000454020.1
|
HJURP
|
Holliday junction recognition protein |
| chr11_+_33563618 | 0.04 |
ENST00000526400.1
|
KIAA1549L
|
KIAA1549-like |
| chr1_-_220219775 | 0.04 |
ENST00000609181.1
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr10_+_75936444 | 0.04 |
ENST00000372734.3
ENST00000541550.1 |
ADK
|
adenosine kinase |
| chr12_-_55042140 | 0.04 |
ENST00000293371.6
ENST00000456047.2 |
DCD
|
dermcidin |
| chr7_-_103086598 | 0.04 |
ENST00000339444.6
ENST00000356767.4 ENST00000393735.2 ENST00000306312.3 ENST00000432958.2 |
SLC26A5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr8_+_92114060 | 0.04 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr16_-_31228680 | 0.04 |
ENST00000302964.3
|
PYDC1
|
PYD (pyrin domain) containing 1 |
| chr17_+_8339340 | 0.04 |
ENST00000580012.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr11_-_47870019 | 0.04 |
ENST00000378460.2
|
NUP160
|
nucleoporin 160kDa |
| chr12_-_123459105 | 0.04 |
ENST00000543935.1
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chrX_-_106243451 | 0.04 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr3_-_10334585 | 0.04 |
ENST00000430179.1
ENST00000449238.2 ENST00000437422.2 ENST00000287656.7 ENST00000457360.1 ENST00000439975.2 ENST00000446937.2 |
GHRL
|
ghrelin/obestatin prepropeptide |
| chr1_-_89458604 | 0.04 |
ENST00000260508.4
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chrX_-_106243294 | 0.04 |
ENST00000255495.7
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr13_-_95248511 | 0.04 |
ENST00000261296.5
|
TGDS
|
TDP-glucose 4,6-dehydratase |
| chr2_-_10978103 | 0.04 |
ENST00000404824.2
|
PDIA6
|
protein disulfide isomerase family A, member 6 |
| chr2_-_69664586 | 0.04 |
ENST00000303698.3
ENST00000394305.1 ENST00000410022.2 |
NFU1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr3_+_93698974 | 0.04 |
ENST00000535334.1
ENST00000478400.1 ENST00000303097.7 ENST00000394222.3 ENST00000471138.1 ENST00000539730.1 |
ARL13B
|
ADP-ribosylation factor-like 13B |
| chr11_+_34645791 | 0.04 |
ENST00000529527.1
ENST00000531728.1 ENST00000525253.1 |
EHF
|
ets homologous factor |
| chr17_+_75452447 | 0.04 |
ENST00000591472.2
|
SEPT9
|
septin 9 |
| chr8_-_86132568 | 0.04 |
ENST00000321777.5
ENST00000458398.2 ENST00000431163.2 ENST00000417663.2 ENST00000524353.1 ENST00000421308.2 ENST00000545322.1 ENST00000518562.1 |
C8orf59
|
chromosome 8 open reading frame 59 |
| chr1_-_236030216 | 0.04 |
ENST00000389794.3
ENST00000389793.2 |
LYST
|
lysosomal trafficking regulator |
| chr7_+_2393714 | 0.04 |
ENST00000431643.1
|
EIF3B
|
eukaryotic translation initiation factor 3, subunit B |
| chr3_-_179322416 | 0.04 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.2 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.1 | GO:0051808 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.1 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0051466 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.1 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.0 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.0 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.0 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.0 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 0.0 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.0 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.0 | 0.1 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.0 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:1901911 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.0 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.0 | GO:0038162 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.0 | GO:0005985 | sucrose metabolic process(GO:0005985) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.0 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.0 | GO:0048007 | synaptic vesicle recycling via endosome(GO:0036466) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.0 | 0.0 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.0 | 0.0 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.0 | 0.0 | GO:0075341 | host cell PML body(GO:0075341) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.0 | 0.1 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.0 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.0 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.0 | 0.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0052841 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.0 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |