Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
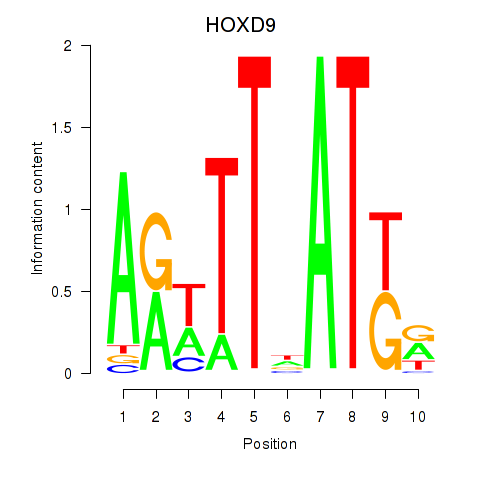
Results for HOXD9
Z-value: 0.40
Transcription factors associated with HOXD9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD9
|
ENSG00000128709.10 | homeobox D9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD9 | hg19_v2_chr2_+_176987088_176987088 | 0.19 | 8.1e-01 | Click! |
Activity profile of HOXD9 motif
Sorted Z-values of HOXD9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91546926 | 0.18 |
ENST00000550758.1
|
DCN
|
decorin |
| chr9_-_75488984 | 0.16 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr6_-_138833630 | 0.15 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr3_+_195413160 | 0.15 |
ENST00000599448.1
|
LINC00969
|
long intergenic non-protein coding RNA 969 |
| chr4_+_39640787 | 0.15 |
ENST00000532680.1
|
RP11-539G18.2
|
RP11-539G18.2 |
| chr4_-_76944621 | 0.15 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr13_-_44735393 | 0.15 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr3_-_195310802 | 0.14 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr1_+_95616933 | 0.14 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr5_-_65018834 | 0.13 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr7_-_130353553 | 0.12 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr19_-_54618650 | 0.12 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr12_-_11002063 | 0.12 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr2_-_191115229 | 0.12 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr17_-_46690839 | 0.11 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr1_-_238108575 | 0.11 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr5_-_119669160 | 0.10 |
ENST00000514240.1
|
CTC-552D5.1
|
CTC-552D5.1 |
| chr9_-_138391692 | 0.09 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr5_+_135496675 | 0.09 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr10_-_94301107 | 0.09 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr17_+_58018269 | 0.09 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr17_+_61151306 | 0.09 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr15_+_49715449 | 0.09 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr6_-_10115007 | 0.09 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr6_-_31632962 | 0.08 |
ENST00000456540.1
ENST00000445768.1 |
GPANK1
|
G patch domain and ankyrin repeats 1 |
| chr5_+_177435986 | 0.08 |
ENST00000398106.2
|
FAM153C
|
family with sequence similarity 153, member C |
| chr11_+_112047087 | 0.08 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr17_+_44679808 | 0.08 |
ENST00000571172.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr12_-_8765446 | 0.08 |
ENST00000537228.1
ENST00000229335.6 |
AICDA
|
activation-induced cytidine deaminase |
| chr3_+_107364769 | 0.08 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr1_+_45140400 | 0.08 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr5_-_78808617 | 0.08 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr4_+_169633310 | 0.07 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_+_75728419 | 0.07 |
ENST00000378695.4
ENST00000312442.2 |
GLIPR1L1
|
GLI pathogenesis-related 1 like 1 |
| chrX_+_71401570 | 0.07 |
ENST00000496835.2
ENST00000446576.1 |
PIN4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr2_-_55647057 | 0.07 |
ENST00000436346.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr12_-_15104040 | 0.07 |
ENST00000541644.1
ENST00000545895.1 |
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr12_-_95510743 | 0.07 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr18_+_21452804 | 0.07 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr2_+_109223595 | 0.07 |
ENST00000410093.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr8_-_25281747 | 0.07 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr4_-_144826682 | 0.07 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr7_-_107880508 | 0.07 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr3_+_182511266 | 0.06 |
ENST00000323116.5
ENST00000493826.1 |
ATP11B
|
ATPase, class VI, type 11B |
| chr4_+_186990298 | 0.06 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr5_+_158654712 | 0.06 |
ENST00000520323.1
|
CTB-11I22.2
|
CTB-11I22.2 |
| chr2_+_109237717 | 0.06 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr5_+_125967372 | 0.06 |
ENST00000357147.3
|
C5orf48
|
chromosome 5 open reading frame 48 |
| chr1_+_219347203 | 0.06 |
ENST00000366927.3
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr6_-_131321863 | 0.06 |
ENST00000528282.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr3_-_182703688 | 0.06 |
ENST00000466812.1
ENST00000487822.1 ENST00000460412.1 ENST00000469954.1 |
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr3_-_107777208 | 0.06 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr19_-_5903714 | 0.06 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr1_-_26231589 | 0.06 |
ENST00000374291.1
|
STMN1
|
stathmin 1 |
| chr12_-_21927736 | 0.06 |
ENST00000240662.2
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr2_+_191334212 | 0.06 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr13_-_81801115 | 0.06 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr7_-_92146729 | 0.06 |
ENST00000541751.1
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr12_-_122879969 | 0.06 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr9_+_70856397 | 0.06 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr12_-_118628350 | 0.05 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr12_-_10601963 | 0.05 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr12_+_41136144 | 0.05 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr7_-_35013217 | 0.05 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr19_+_45843994 | 0.05 |
ENST00000391946.2
|
KLC3
|
kinesin light chain 3 |
| chr19_+_52800410 | 0.05 |
ENST00000595962.1
ENST00000598016.1 ENST00000334564.7 ENST00000490272.1 |
ZNF480
|
zinc finger protein 480 |
| chr1_-_95391315 | 0.05 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr4_-_84035905 | 0.05 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr18_-_57027194 | 0.05 |
ENST00000251047.5
|
LMAN1
|
lectin, mannose-binding, 1 |
| chr1_-_100643765 | 0.05 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr3_-_27498235 | 0.05 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr2_-_55646957 | 0.05 |
ENST00000263630.8
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr3_+_107364683 | 0.05 |
ENST00000413213.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr2_-_170430277 | 0.05 |
ENST00000438035.1
ENST00000453929.2 |
FASTKD1
|
FAST kinase domains 1 |
| chr14_-_31856397 | 0.05 |
ENST00000538864.2
ENST00000550366.1 |
HEATR5A
|
HEAT repeat containing 5A |
| chr15_+_49913175 | 0.05 |
ENST00000403028.3
|
DTWD1
|
DTW domain containing 1 |
| chr14_+_20215587 | 0.04 |
ENST00000331723.1
|
OR4Q3
|
olfactory receptor, family 4, subfamily Q, member 3 |
| chr10_+_115511213 | 0.04 |
ENST00000361048.1
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr10_-_31146615 | 0.04 |
ENST00000444692.2
|
ZNF438
|
zinc finger protein 438 |
| chr1_-_213020991 | 0.04 |
ENST00000332912.3
|
C1orf227
|
chromosome 1 open reading frame 227 |
| chrX_-_10645724 | 0.04 |
ENST00000413894.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr7_+_77469439 | 0.04 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr12_-_118628315 | 0.04 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr3_+_107318157 | 0.04 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr17_-_67224812 | 0.04 |
ENST00000423818.2
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10 |
| chr22_-_19466643 | 0.04 |
ENST00000474226.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr4_-_100065440 | 0.04 |
ENST00000508393.1
ENST00000265512.7 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr2_-_170430366 | 0.04 |
ENST00000453153.2
ENST00000445210.1 |
FASTKD1
|
FAST kinase domains 1 |
| chr1_-_248005254 | 0.04 |
ENST00000355784.2
|
OR11L1
|
olfactory receptor, family 11, subfamily L, member 1 |
| chr10_-_13523073 | 0.04 |
ENST00000440282.1
|
BEND7
|
BEN domain containing 7 |
| chr4_+_74606223 | 0.04 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr4_-_144940477 | 0.04 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr7_+_117864708 | 0.04 |
ENST00000357099.4
ENST00000265224.4 ENST00000486422.1 ENST00000417525.1 |
ANKRD7
|
ankyrin repeat domain 7 |
| chr18_+_3466248 | 0.04 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr3_-_116163830 | 0.04 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr4_-_120225600 | 0.04 |
ENST00000399075.4
|
C4orf3
|
chromosome 4 open reading frame 3 |
| chr11_-_16430399 | 0.04 |
ENST00000528252.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr6_+_149539767 | 0.04 |
ENST00000606202.1
ENST00000536230.1 ENST00000445901.1 |
TAB2
RP1-111D6.3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr20_+_9146969 | 0.04 |
ENST00000416836.1
|
PLCB4
|
phospholipase C, beta 4 |
| chrX_+_123097014 | 0.04 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr2_+_135809835 | 0.04 |
ENST00000264158.8
ENST00000539493.1 ENST00000442034.1 ENST00000425393.1 |
RAB3GAP1
|
RAB3 GTPase activating protein subunit 1 (catalytic) |
| chr19_-_56826157 | 0.04 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr14_+_65878650 | 0.03 |
ENST00000555559.1
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr1_-_48937838 | 0.03 |
ENST00000371847.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr2_+_28824786 | 0.03 |
ENST00000541605.1
|
PLB1
|
phospholipase B1 |
| chr12_+_19358228 | 0.03 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr1_+_81771806 | 0.03 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr10_+_104613980 | 0.03 |
ENST00000339834.5
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr14_-_38028689 | 0.03 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr8_-_66701319 | 0.03 |
ENST00000379419.4
|
PDE7A
|
phosphodiesterase 7A |
| chr2_-_128284020 | 0.03 |
ENST00000295321.4
ENST00000455721.2 |
IWS1
|
IWS1 homolog (S. cerevisiae) |
| chr5_-_96518907 | 0.03 |
ENST00000508447.1
ENST00000283109.3 |
RIOK2
|
RIO kinase 2 |
| chr15_-_75748115 | 0.03 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr20_+_19870167 | 0.03 |
ENST00000440354.2
|
RIN2
|
Ras and Rab interactor 2 |
| chr7_-_50633078 | 0.03 |
ENST00000444124.2
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr11_+_107461804 | 0.03 |
ENST00000531234.1
|
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr15_-_75748143 | 0.03 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr8_-_102181718 | 0.03 |
ENST00000565617.1
|
KB-1460A1.5
|
KB-1460A1.5 |
| chr13_+_51483814 | 0.03 |
ENST00000336617.3
ENST00000422660.1 |
RNASEH2B
|
ribonuclease H2, subunit B |
| chr1_-_236046872 | 0.03 |
ENST00000536965.1
|
LYST
|
lysosomal trafficking regulator |
| chr22_+_19118321 | 0.03 |
ENST00000399635.2
|
TSSK2
|
testis-specific serine kinase 2 |
| chr11_-_8986474 | 0.03 |
ENST00000525069.1
|
TMEM9B
|
TMEM9 domain family, member B |
| chr1_-_244006528 | 0.03 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr11_-_102401469 | 0.03 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr8_+_86089619 | 0.03 |
ENST00000256117.5
ENST00000416274.2 |
E2F5
|
E2F transcription factor 5, p130-binding |
| chr3_-_139396853 | 0.03 |
ENST00000406164.1
ENST00000406824.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr8_-_117886955 | 0.03 |
ENST00000297338.2
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr13_+_33160553 | 0.03 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr4_-_185395191 | 0.03 |
ENST00000510814.1
ENST00000507523.1 ENST00000506230.1 |
IRF2
|
interferon regulatory factor 2 |
| chr14_+_62164340 | 0.03 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr11_-_104905840 | 0.03 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr2_-_175712270 | 0.03 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr2_+_210444748 | 0.02 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr4_+_37455536 | 0.02 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr10_+_115614370 | 0.02 |
ENST00000369301.3
|
NHLRC2
|
NHL repeat containing 2 |
| chr15_+_57998923 | 0.02 |
ENST00000380557.4
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr17_+_57233087 | 0.02 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr9_+_20927764 | 0.02 |
ENST00000603044.1
ENST00000604254.1 |
FOCAD
|
focadhesin |
| chr2_-_225434538 | 0.02 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr7_+_16793160 | 0.02 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr1_+_44870866 | 0.02 |
ENST00000355387.2
ENST00000361799.2 |
RNF220
|
ring finger protein 220 |
| chr1_+_45140360 | 0.02 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr9_+_96846740 | 0.02 |
ENST00000288976.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr16_-_21442874 | 0.02 |
ENST00000534903.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr9_-_123812542 | 0.02 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr1_+_78383813 | 0.02 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr6_+_24775153 | 0.02 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr19_-_54619006 | 0.02 |
ENST00000391759.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr1_+_82165350 | 0.02 |
ENST00000359929.3
|
LPHN2
|
latrophilin 2 |
| chrX_-_68385354 | 0.02 |
ENST00000361478.1
|
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr1_+_197881592 | 0.02 |
ENST00000367391.1
ENST00000367390.3 |
LHX9
|
LIM homeobox 9 |
| chr15_+_52155001 | 0.02 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr12_-_118797475 | 0.02 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr1_-_197036364 | 0.02 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr6_+_1312675 | 0.02 |
ENST00000296839.2
|
FOXQ1
|
forkhead box Q1 |
| chrX_-_68385274 | 0.02 |
ENST00000374584.3
ENST00000590146.1 |
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr14_+_20528204 | 0.02 |
ENST00000315683.1
|
OR4L1
|
olfactory receptor, family 4, subfamily L, member 1 |
| chr5_+_61874562 | 0.02 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr15_-_49913126 | 0.02 |
ENST00000561064.1
ENST00000299338.6 |
FAM227B
|
family with sequence similarity 227, member B |
| chr1_-_48937821 | 0.02 |
ENST00000396199.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr5_+_112043186 | 0.02 |
ENST00000509732.1
ENST00000457016.1 ENST00000507379.1 |
APC
|
adenomatous polyposis coli |
| chr1_-_241799232 | 0.02 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr7_-_107204706 | 0.02 |
ENST00000393603.2
|
COG5
|
component of oligomeric golgi complex 5 |
| chr3_+_158991025 | 0.02 |
ENST00000337808.6
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr6_-_52859968 | 0.02 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr7_-_5998714 | 0.02 |
ENST00000539903.1
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr4_+_71263599 | 0.02 |
ENST00000399575.2
|
PROL1
|
proline rich, lacrimal 1 |
| chr1_+_109255279 | 0.02 |
ENST00000370017.3
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr20_-_20693131 | 0.02 |
ENST00000202677.7
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr3_-_139396801 | 0.02 |
ENST00000413939.2
ENST00000339837.5 ENST00000512391.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr12_-_30887948 | 0.02 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chr11_+_69924639 | 0.02 |
ENST00000538023.1
ENST00000398543.2 |
ANO1
|
anoctamin 1, calcium activated chloride channel |
| chr4_-_84035868 | 0.02 |
ENST00000426923.2
ENST00000509973.1 |
PLAC8
|
placenta-specific 8 |
| chrY_+_14958970 | 0.02 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr7_-_6866401 | 0.01 |
ENST00000316731.8
|
CCZ1B
|
CCZ1 vacuolar protein trafficking and biogenesis associated homolog B (S. cerevisiae) |
| chrX_+_118602363 | 0.01 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr14_+_81421355 | 0.01 |
ENST00000541158.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chr2_+_210517895 | 0.01 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr8_-_90993869 | 0.01 |
ENST00000517772.1
|
NBN
|
nibrin |
| chr19_-_19739321 | 0.01 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr4_-_146859787 | 0.01 |
ENST00000508784.1
|
ZNF827
|
zinc finger protein 827 |
| chr16_+_22518495 | 0.01 |
ENST00000541154.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr10_+_98064085 | 0.01 |
ENST00000419175.1
ENST00000371174.2 |
DNTT
|
DNA nucleotidylexotransferase |
| chr11_-_128457446 | 0.01 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr10_-_115614127 | 0.01 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr6_+_10528560 | 0.01 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr17_-_54893250 | 0.01 |
ENST00000397862.2
|
C17orf67
|
chromosome 17 open reading frame 67 |
| chrX_-_130423240 | 0.01 |
ENST00000370910.1
ENST00000370901.4 |
IGSF1
|
immunoglobulin superfamily, member 1 |
| chrX_-_130423200 | 0.01 |
ENST00000361420.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr15_+_76352178 | 0.01 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr6_-_39399087 | 0.01 |
ENST00000229913.5
ENST00000541946.1 ENST00000394362.1 |
KIF6
|
kinesin family member 6 |
| chr20_-_39928756 | 0.01 |
ENST00000432768.2
|
ZHX3
|
zinc fingers and homeoboxes 3 |
| chr19_-_53466095 | 0.01 |
ENST00000391786.2
ENST00000434371.2 ENST00000357666.4 ENST00000438970.2 ENST00000270457.4 ENST00000535506.1 ENST00000444460.2 ENST00000457013.2 |
ZNF816
|
zinc finger protein 816 |
| chr11_+_69924679 | 0.01 |
ENST00000531604.1
|
ANO1
|
anoctamin 1, calcium activated chloride channel |
| chr1_+_50459990 | 0.01 |
ENST00000448346.1
|
AL645730.2
|
AL645730.2 |
| chr12_-_10007448 | 0.01 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr2_-_136288113 | 0.01 |
ENST00000401392.1
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr3_+_169539710 | 0.01 |
ENST00000340806.6
|
LRRIQ4
|
leucine-rich repeats and IQ motif containing 4 |
| chr10_-_99030395 | 0.01 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr14_+_92789498 | 0.01 |
ENST00000531433.1
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
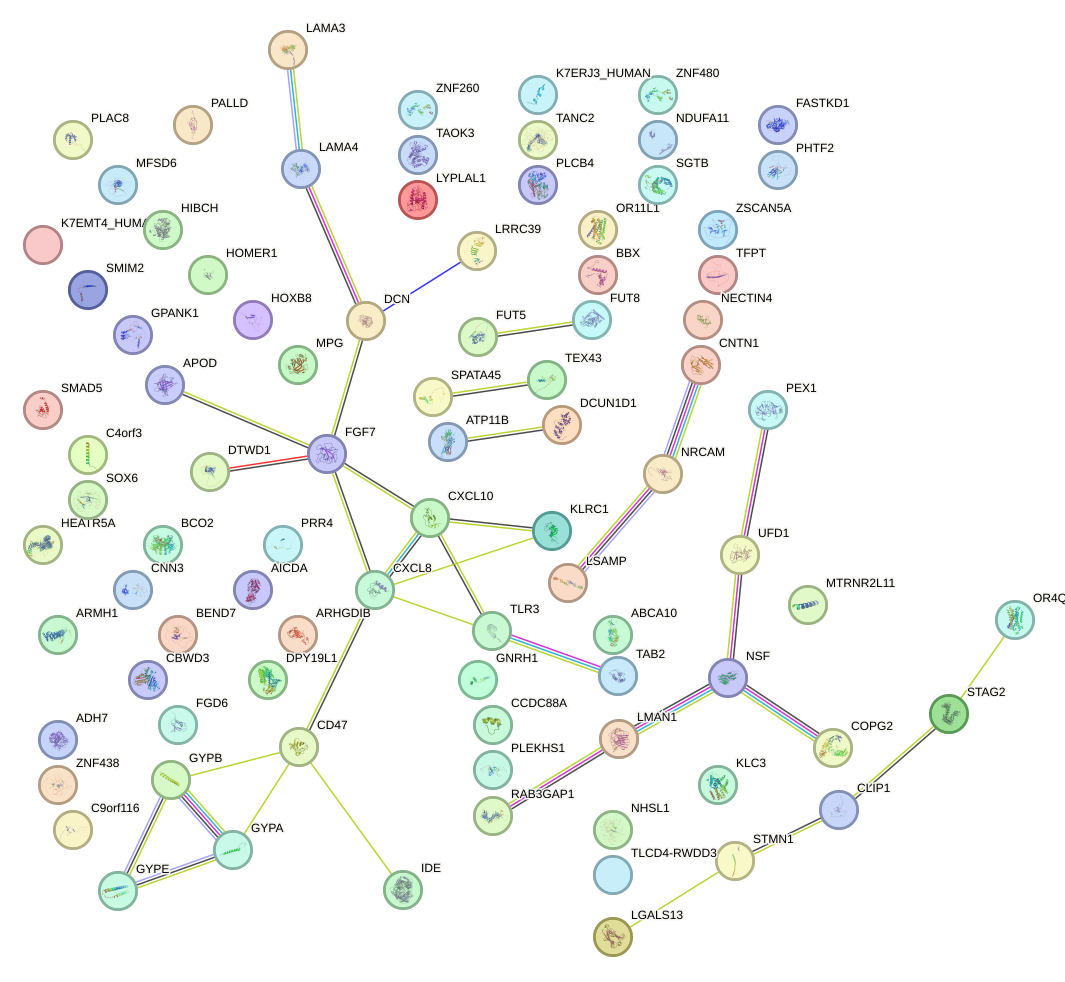
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.0 | GO:1903371 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.0 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.0 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) isoquinoline alkaloid metabolic process(GO:0033076) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.0 | GO:0035276 | all-trans retinal binding(GO:0005503) ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |