Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
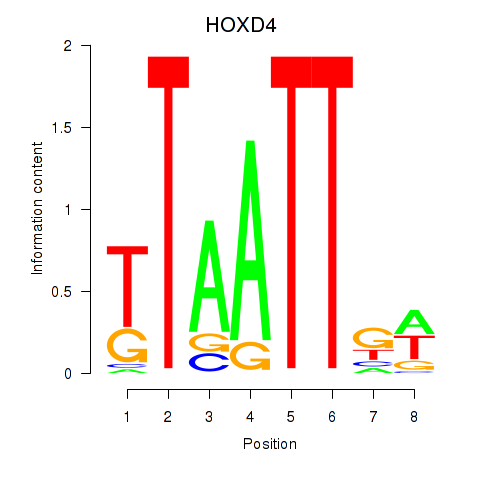
Results for HOXD4
Z-value: 0.44
Transcription factors associated with HOXD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD4
|
ENSG00000170166.5 | homeobox D4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD4 | hg19_v2_chr2_+_177015950_177015950 | -0.39 | 6.1e-01 | Click! |
Activity profile of HOXD4 motif
Sorted Z-values of HOXD4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_26250835 | 0.25 |
ENST00000446824.2
|
HIST1H3F
|
histone cluster 1, H3f |
| chr18_+_68002675 | 0.24 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr2_-_99871570 | 0.22 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr2_+_90198535 | 0.20 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr3_-_178103144 | 0.19 |
ENST00000417383.1
ENST00000418585.1 ENST00000411727.1 ENST00000439810.1 |
RP11-33A14.1
|
RP11-33A14.1 |
| chr3_-_196987309 | 0.18 |
ENST00000453607.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr1_+_149754227 | 0.18 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr1_-_238108575 | 0.18 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr15_-_94614049 | 0.18 |
ENST00000556447.1
ENST00000555772.1 |
CTD-3049M7.1
|
CTD-3049M7.1 |
| chr9_+_97562440 | 0.16 |
ENST00000395357.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr11_+_128563652 | 0.16 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr4_-_89442940 | 0.14 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr17_+_9479944 | 0.12 |
ENST00000396219.3
ENST00000352665.5 |
WDR16
|
WD repeat domain 16 |
| chr5_-_150727111 | 0.12 |
ENST00000335244.4
ENST00000521967.1 |
SLC36A2
|
solute carrier family 36 (proton/amino acid symporter), member 2 |
| chr2_+_196313239 | 0.12 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr8_+_107738343 | 0.11 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr7_-_137028534 | 0.11 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr12_+_28605426 | 0.11 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr9_-_128246769 | 0.11 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr4_+_124571409 | 0.11 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr1_-_234831899 | 0.10 |
ENST00000442382.1
|
RP4-781K5.6
|
RP4-781K5.6 |
| chr7_-_137028498 | 0.10 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr3_-_4927447 | 0.10 |
ENST00000449914.1
|
AC018816.3
|
Uncharacterized protein |
| chr1_+_22138758 | 0.10 |
ENST00000344642.2
ENST00000543870.1 |
LDLRAD2
|
low density lipoprotein receptor class A domain containing 2 |
| chr21_-_27423339 | 0.09 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr3_-_11623804 | 0.09 |
ENST00000451674.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr14_-_54425475 | 0.09 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr4_-_105416039 | 0.09 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr12_-_118628315 | 0.09 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr3_+_69812877 | 0.08 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr2_-_190927447 | 0.08 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr5_+_101569696 | 0.08 |
ENST00000597120.1
|
AC008948.1
|
AC008948.1 |
| chr2_+_176981307 | 0.08 |
ENST00000249501.4
|
HOXD10
|
homeobox D10 |
| chr11_+_112041253 | 0.07 |
ENST00000532612.1
|
AP002884.3
|
AP002884.3 |
| chr6_+_130339710 | 0.07 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr2_+_33701684 | 0.07 |
ENST00000442390.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr16_-_29910853 | 0.07 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr2_-_99279928 | 0.07 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr3_+_140981456 | 0.07 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr5_+_102200948 | 0.07 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr18_+_29027696 | 0.07 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr2_+_179318295 | 0.06 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr10_+_69869237 | 0.06 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr8_-_27941380 | 0.06 |
ENST00000413272.2
ENST00000341513.6 |
NUGGC
|
nuclear GTPase, germinal center associated |
| chr2_-_228497888 | 0.06 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr8_+_24151553 | 0.06 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr11_-_36310958 | 0.06 |
ENST00000532705.1
ENST00000263401.5 ENST00000452374.2 |
COMMD9
|
COMM domain containing 9 |
| chr7_+_135611542 | 0.06 |
ENST00000416501.1
|
AC015987.2
|
AC015987.2 |
| chr19_-_14064114 | 0.06 |
ENST00000585607.1
ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1
|
podocan-like 1 |
| chr1_+_67632083 | 0.05 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr12_-_112279694 | 0.05 |
ENST00000443596.1
ENST00000442119.1 |
MAPKAPK5-AS1
|
MAPKAPK5 antisense RNA 1 |
| chr11_-_31531121 | 0.05 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr15_+_84115868 | 0.05 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr5_-_10761206 | 0.05 |
ENST00000432074.2
ENST00000230895.6 |
DAP
|
death-associated protein |
| chrX_-_92928557 | 0.05 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr16_+_84801852 | 0.05 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr11_-_64684672 | 0.05 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr1_+_214161854 | 0.04 |
ENST00000435016.1
|
PROX1
|
prospero homeobox 1 |
| chrX_-_110655306 | 0.04 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr13_+_24144509 | 0.04 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr19_+_50016610 | 0.04 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr19_+_48828788 | 0.04 |
ENST00000594198.1
ENST00000597279.1 ENST00000593437.1 |
EMP3
|
epithelial membrane protein 3 |
| chr1_+_202385953 | 0.04 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr14_-_24911868 | 0.04 |
ENST00000554698.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr14_+_23067146 | 0.04 |
ENST00000428304.2
|
ABHD4
|
abhydrolase domain containing 4 |
| chr1_+_144146808 | 0.04 |
ENST00000369190.5
ENST00000412624.2 ENST00000369365.3 |
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr3_+_142342228 | 0.04 |
ENST00000337777.3
|
PLS1
|
plastin 1 |
| chr8_-_124279627 | 0.04 |
ENST00000357082.4
|
ZHX1-C8ORF76
|
ZHX1-C8ORF76 readthrough |
| chr3_+_51575596 | 0.04 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr3_+_63638372 | 0.04 |
ENST00000496807.1
|
SNTN
|
sentan, cilia apical structure protein |
| chr15_+_36887069 | 0.04 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr10_+_18629628 | 0.04 |
ENST00000377329.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr1_+_186798073 | 0.04 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr2_-_27579842 | 0.04 |
ENST00000423998.1
ENST00000264720.3 |
GTF3C2
|
general transcription factor IIIC, polypeptide 2, beta 110kDa |
| chr14_+_23067166 | 0.04 |
ENST00000216327.6
ENST00000542041.1 |
ABHD4
|
abhydrolase domain containing 4 |
| chr12_+_16109519 | 0.03 |
ENST00000526530.1
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr1_-_101360331 | 0.03 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr17_-_64225508 | 0.03 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr22_+_44427230 | 0.03 |
ENST00000444029.1
|
PARVB
|
parvin, beta |
| chr17_+_47448102 | 0.03 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr14_-_99947168 | 0.03 |
ENST00000331768.5
|
SETD3
|
SET domain containing 3 |
| chr6_+_142468361 | 0.03 |
ENST00000367630.4
|
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr4_-_130014532 | 0.03 |
ENST00000506368.1
ENST00000439369.2 ENST00000503215.1 |
SCLT1
|
sodium channel and clathrin linker 1 |
| chr14_+_62164340 | 0.03 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr18_+_22040593 | 0.03 |
ENST00000256906.4
|
HRH4
|
histamine receptor H4 |
| chr1_+_159750776 | 0.03 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr1_-_231560790 | 0.03 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr14_+_61449076 | 0.03 |
ENST00000526105.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr1_-_235814048 | 0.03 |
ENST00000450593.1
ENST00000366598.4 |
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4 |
| chr7_+_16793160 | 0.03 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr14_+_78174414 | 0.03 |
ENST00000557342.1
ENST00000238688.5 ENST00000557623.1 ENST00000557431.1 ENST00000556831.1 ENST00000556375.1 ENST00000553981.1 |
SLIRP
|
SRA stem-loop interacting RNA binding protein |
| chr8_-_40200877 | 0.03 |
ENST00000521030.1
|
CTA-392C11.1
|
CTA-392C11.1 |
| chr5_-_78809950 | 0.03 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chr2_-_31361543 | 0.03 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr5_+_52083730 | 0.03 |
ENST00000282588.6
ENST00000274311.2 |
ITGA1
PELO
|
integrin, alpha 1 pelota homolog (Drosophila) |
| chr5_+_141346385 | 0.03 |
ENST00000513019.1
ENST00000356143.1 |
RNF14
|
ring finger protein 14 |
| chr16_+_103816 | 0.03 |
ENST00000383018.3
ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr6_-_44281043 | 0.02 |
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr5_-_179499108 | 0.02 |
ENST00000521389.1
|
RNF130
|
ring finger protein 130 |
| chr12_-_39734783 | 0.02 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr20_+_18447771 | 0.02 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr22_-_29107919 | 0.02 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr22_-_30642728 | 0.02 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr17_-_41132410 | 0.02 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr8_-_102803163 | 0.02 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr3_-_18480260 | 0.02 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr17_+_41132564 | 0.02 |
ENST00000361677.1
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr5_+_68860949 | 0.02 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr15_+_80364901 | 0.02 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr3_+_97483572 | 0.02 |
ENST00000335979.2
ENST00000394206.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr2_+_166095898 | 0.02 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr3_+_15045419 | 0.02 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr1_+_109256067 | 0.02 |
ENST00000271311.2
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr1_-_28384598 | 0.02 |
ENST00000373864.1
|
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chr10_-_17171817 | 0.02 |
ENST00000377833.4
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr4_-_39979576 | 0.02 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr2_+_234826016 | 0.02 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr1_+_109265033 | 0.02 |
ENST00000445274.1
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr8_-_42698433 | 0.02 |
ENST00000345117.2
ENST00000254250.3 |
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr12_+_112279782 | 0.02 |
ENST00000550735.2
|
MAPKAPK5
|
mitogen-activated protein kinase-activated protein kinase 5 |
| chr4_+_71019903 | 0.02 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr15_+_48483736 | 0.02 |
ENST00000417307.2
ENST00000559641.1 |
CTXN2
SLC12A1
|
cortexin 2 solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr13_-_24007815 | 0.02 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr18_+_32556892 | 0.02 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr7_+_112063192 | 0.02 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr1_+_84630645 | 0.01 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr3_+_158787041 | 0.01 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr15_+_55700741 | 0.01 |
ENST00000569691.1
|
C15orf65
|
chromosome 15 open reading frame 65 |
| chr3_-_137851220 | 0.01 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr15_-_56209306 | 0.01 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr5_+_140762268 | 0.01 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr5_+_66300446 | 0.01 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr2_+_234590556 | 0.01 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr2_-_165424973 | 0.01 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr1_+_42846443 | 0.01 |
ENST00000410070.2
ENST00000431473.3 |
RIMKLA
|
ribosomal modification protein rimK-like family member A |
| chr1_+_197237352 | 0.01 |
ENST00000538660.1
ENST00000367400.3 ENST00000367399.2 |
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr2_+_171034646 | 0.01 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr11_+_101983176 | 0.01 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr19_-_5903714 | 0.01 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr6_+_53659746 | 0.01 |
ENST00000370888.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chr12_+_12938541 | 0.01 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr12_+_26348246 | 0.01 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr17_-_26733179 | 0.01 |
ENST00000440501.1
ENST00000321666.5 |
SLC46A1
|
solute carrier family 46 (folate transporter), member 1 |
| chr17_-_7218631 | 0.01 |
ENST00000577040.2
ENST00000389167.5 ENST00000391950.3 |
GPS2
|
G protein pathway suppressor 2 |
| chr6_-_53013620 | 0.01 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr17_-_78450398 | 0.01 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr3_-_149293990 | 0.01 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr18_-_74839891 | 0.01 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr4_-_186570679 | 0.01 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_-_131790550 | 0.01 |
ENST00000372554.4
ENST00000372564.3 |
SH3GLB2
|
SH3-domain GRB2-like endophilin B2 |
| chr19_+_30433110 | 0.01 |
ENST00000542441.2
ENST00000392271.1 |
URI1
|
URI1, prefoldin-like chaperone |
| chr4_-_152149033 | 0.01 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr2_-_166060571 | 0.01 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr4_-_138453606 | 0.01 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr14_-_24911971 | 0.01 |
ENST00000555365.1
ENST00000399395.3 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr2_-_176046391 | 0.01 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr14_+_29236269 | 0.01 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr1_-_101360374 | 0.01 |
ENST00000535414.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr4_-_46996424 | 0.01 |
ENST00000264318.3
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chr1_-_9953295 | 0.01 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr1_-_190446759 | 0.01 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr5_+_179921344 | 0.01 |
ENST00000261951.4
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr2_+_32502952 | 0.01 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr19_+_48828582 | 0.01 |
ENST00000270221.6
ENST00000596315.1 |
EMP3
|
epithelial membrane protein 3 |
| chr8_-_116504448 | 0.01 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr10_-_50970322 | 0.01 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr14_+_90863327 | 0.01 |
ENST00000356978.4
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr5_-_110848189 | 0.00 |
ENST00000296632.3
ENST00000512160.1 ENST00000509887.1 |
STARD4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr2_+_110656005 | 0.00 |
ENST00000437679.2
|
LIMS3
|
LIM and senescent cell antigen-like domains 3 |
| chr4_+_74347400 | 0.00 |
ENST00000226355.3
|
AFM
|
afamin |
| chr11_-_89224638 | 0.00 |
ENST00000535633.1
ENST00000263317.4 |
NOX4
|
NADPH oxidase 4 |
| chr17_-_60142609 | 0.00 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chrX_-_13835147 | 0.00 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr15_-_64386120 | 0.00 |
ENST00000300030.3
|
FAM96A
|
family with sequence similarity 96, member A |
| chr1_-_151762900 | 0.00 |
ENST00000440583.2
|
TDRKH
|
tudor and KH domain containing |
| chr12_+_9066472 | 0.00 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr15_-_64385981 | 0.00 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr2_-_225434538 | 0.00 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr4_+_169418195 | 0.00 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_-_92952433 | 0.00 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr16_+_24549014 | 0.00 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr3_-_4508925 | 0.00 |
ENST00000534863.1
ENST00000383843.5 ENST00000458465.2 ENST00000405420.2 ENST00000272902.5 |
SUMF1
|
sulfatase modifying factor 1 |
| chr5_+_140743859 | 0.00 |
ENST00000518069.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr10_+_48189612 | 0.00 |
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:0003277 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.0 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0031179 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.0 | 0.0 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0015193 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.1 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.0 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |