Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
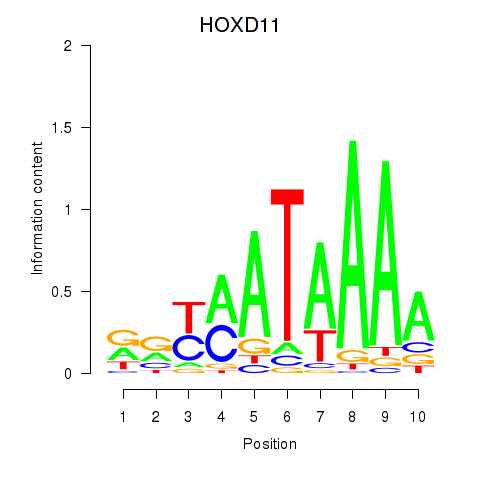
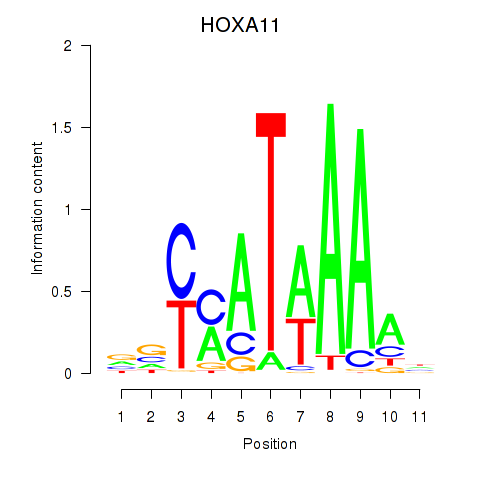
Results for HOXD11_HOXA11
Z-value: 0.56
Transcription factors associated with HOXD11_HOXA11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD11
|
ENSG00000128713.11 | homeobox D11 |
|
HOXA11
|
ENSG00000005073.5 | homeobox A11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD11 | hg19_v2_chr2_+_176972000_176972025 | -0.78 | 2.2e-01 | Click! |
| HOXA11 | hg19_v2_chr7_-_27224842_27224872 | -0.60 | 4.0e-01 | Click! |
Activity profile of HOXD11_HOXA11 motif
Sorted Z-values of HOXD11_HOXA11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_46690839 | 0.49 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr12_+_54384370 | 0.27 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr1_-_160231451 | 0.25 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr14_-_51027838 | 0.23 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr2_+_207024306 | 0.22 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr8_+_52730143 | 0.21 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr2_+_38177575 | 0.20 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr16_-_18466642 | 0.20 |
ENST00000541810.1
ENST00000531453.2 |
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr2_+_109204909 | 0.19 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr14_+_39944025 | 0.18 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr18_+_68002675 | 0.17 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr5_-_145562147 | 0.17 |
ENST00000545646.1
ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS
|
leucyl-tRNA synthetase |
| chr1_+_28527070 | 0.17 |
ENST00000596102.1
|
AL353354.2
|
AL353354.2 |
| chr2_+_200472779 | 0.17 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chr2_+_64073187 | 0.17 |
ENST00000491621.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr15_+_35270552 | 0.16 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr4_+_39046615 | 0.16 |
ENST00000261425.3
ENST00000508137.2 |
KLHL5
|
kelch-like family member 5 |
| chr18_+_3252265 | 0.16 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr10_-_115904361 | 0.16 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr9_-_16705069 | 0.15 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr1_+_62439037 | 0.15 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr14_+_52456327 | 0.14 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr9_-_21305312 | 0.14 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr8_+_66955648 | 0.14 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr6_-_13621126 | 0.13 |
ENST00000600057.1
|
AL441883.1
|
Uncharacterized protein |
| chr1_+_229440129 | 0.13 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr4_+_124571409 | 0.13 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr11_-_74204692 | 0.13 |
ENST00000528085.1
|
LIPT2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr4_+_74606223 | 0.13 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr12_-_112443830 | 0.13 |
ENST00000550037.1
ENST00000549425.1 |
TMEM116
|
transmembrane protein 116 |
| chr14_+_93357749 | 0.13 |
ENST00000557689.1
|
RP11-862G15.1
|
RP11-862G15.1 |
| chr14_-_69444957 | 0.13 |
ENST00000556571.1
ENST00000553659.1 ENST00000555616.1 |
ACTN1
|
actinin, alpha 1 |
| chr2_+_37571845 | 0.12 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr5_+_61874562 | 0.12 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr10_+_114709999 | 0.12 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr2_+_109204743 | 0.12 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr13_-_44735393 | 0.12 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr11_+_120039685 | 0.12 |
ENST00000530303.1
ENST00000319763.1 |
AP000679.2
|
Uncharacterized protein |
| chr7_+_108210012 | 0.12 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr12_-_95510743 | 0.11 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr10_-_101673782 | 0.11 |
ENST00000422692.1
|
DNMBP
|
dynamin binding protein |
| chr1_-_154178803 | 0.11 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr2_+_67624430 | 0.11 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr7_-_69062391 | 0.11 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr12_-_91546926 | 0.11 |
ENST00000550758.1
|
DCN
|
decorin |
| chr16_+_15031300 | 0.11 |
ENST00000328085.6
|
NPIPA1
|
nuclear pore complex interacting protein family, member A1 |
| chr17_-_48943706 | 0.11 |
ENST00000499247.2
|
TOB1
|
transducer of ERBB2, 1 |
| chr19_+_30433372 | 0.11 |
ENST00000312051.6
|
URI1
|
URI1, prefoldin-like chaperone |
| chr16_+_16429787 | 0.11 |
ENST00000331436.4
ENST00000541593.1 |
AC138969.4
|
Protein PKD1P1 |
| chr18_-_12656715 | 0.10 |
ENST00000462226.1
ENST00000497844.2 ENST00000309836.5 ENST00000453447.2 |
SPIRE1
|
spire-type actin nucleation factor 1 |
| chr10_+_85933494 | 0.10 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr7_-_112635675 | 0.10 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr5_+_148521454 | 0.10 |
ENST00000508983.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr9_+_103189458 | 0.10 |
ENST00000398977.2
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr4_+_174089951 | 0.10 |
ENST00000512285.1
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr2_-_136678123 | 0.10 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr3_+_15045419 | 0.10 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr1_+_23345943 | 0.10 |
ENST00000400181.4
ENST00000542151.1 |
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr7_-_108209897 | 0.10 |
ENST00000313516.5
|
THAP5
|
THAP domain containing 5 |
| chr3_-_37216055 | 0.10 |
ENST00000336686.4
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr11_+_34663913 | 0.10 |
ENST00000532302.1
|
EHF
|
ets homologous factor |
| chr12_+_86268065 | 0.10 |
ENST00000551529.1
ENST00000256010.6 |
NTS
|
neurotensin |
| chrX_-_37706815 | 0.09 |
ENST00000378578.4
|
DYNLT3
|
dynein, light chain, Tctex-type 3 |
| chr8_+_11225911 | 0.09 |
ENST00000284481.3
|
C8orf12
|
chromosome 8 open reading frame 12 |
| chr4_-_39033963 | 0.09 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr2_-_207023918 | 0.09 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr14_+_62164340 | 0.09 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr10_-_14050522 | 0.09 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr3_-_67705006 | 0.09 |
ENST00000492795.1
ENST00000493112.1 ENST00000307227.5 |
SUCLG2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr2_+_109271481 | 0.09 |
ENST00000542845.1
ENST00000393314.2 |
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr3_-_197686847 | 0.09 |
ENST00000265239.6
|
IQCG
|
IQ motif containing G |
| chr16_+_16472912 | 0.09 |
ENST00000530217.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr1_+_23345930 | 0.09 |
ENST00000356634.3
|
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr8_-_73793975 | 0.09 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr5_+_148521381 | 0.09 |
ENST00000504238.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr12_+_104337515 | 0.09 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr5_-_65017921 | 0.09 |
ENST00000381007.4
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr9_+_137979506 | 0.09 |
ENST00000539529.1
ENST00000392991.4 ENST00000371793.3 |
OLFM1
|
olfactomedin 1 |
| chr17_-_39684550 | 0.09 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chr18_-_54305658 | 0.09 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chr10_+_72194585 | 0.09 |
ENST00000420338.2
|
AC022532.1
|
Uncharacterized protein |
| chr17_+_58018269 | 0.09 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr11_-_67373435 | 0.09 |
ENST00000446232.1
|
C11orf72
|
chromosome 11 open reading frame 72 |
| chr12_-_112443767 | 0.09 |
ENST00000550233.1
ENST00000546962.1 ENST00000550800.2 |
TMEM116
|
transmembrane protein 116 |
| chr4_+_86525299 | 0.09 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr6_-_35109145 | 0.09 |
ENST00000373974.4
ENST00000244645.3 |
TCP11
|
t-complex 11, testis-specific |
| chr7_+_134464414 | 0.08 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr8_-_81083890 | 0.08 |
ENST00000518937.1
|
TPD52
|
tumor protein D52 |
| chr16_+_66613351 | 0.08 |
ENST00000379486.2
ENST00000268595.2 |
CMTM2
|
CKLF-like MARVEL transmembrane domain containing 2 |
| chr14_+_52456193 | 0.08 |
ENST00000261700.3
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr13_+_73632897 | 0.08 |
ENST00000377687.4
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr5_+_67586465 | 0.08 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr10_+_35456444 | 0.08 |
ENST00000361599.4
|
CREM
|
cAMP responsive element modulator |
| chr1_+_154909803 | 0.08 |
ENST00000604546.1
|
RP11-307C12.13
|
RP11-307C12.13 |
| chr2_-_191115229 | 0.08 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr2_-_4703793 | 0.08 |
ENST00000421212.1
ENST00000412134.1 |
AC022311.1
|
AC022311.1 |
| chr9_+_103189536 | 0.08 |
ENST00000374885.1
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr22_+_23248512 | 0.08 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chrX_+_13671225 | 0.08 |
ENST00000545566.1
ENST00000544987.1 ENST00000314720.4 |
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr8_-_101730061 | 0.08 |
ENST00000519100.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr12_-_74686314 | 0.08 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr11_-_125648690 | 0.08 |
ENST00000436890.2
ENST00000358524.3 |
PATE2
|
prostate and testis expressed 2 |
| chr10_-_104001231 | 0.08 |
ENST00000370002.3
|
PITX3
|
paired-like homeodomain 3 |
| chr15_-_80263506 | 0.08 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr11_+_36616044 | 0.08 |
ENST00000334307.5
ENST00000531554.1 ENST00000347206.4 ENST00000534635.1 ENST00000446510.2 ENST00000530697.1 ENST00000527108.1 |
C11orf74
|
chromosome 11 open reading frame 74 |
| chr4_+_56815102 | 0.08 |
ENST00000257287.4
|
CEP135
|
centrosomal protein 135kDa |
| chr5_+_136070614 | 0.08 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr2_-_111230393 | 0.08 |
ENST00000447537.2
ENST00000413601.2 |
LIMS3L
LIMS3L
|
LIM and senescent cell antigen-like-containing domain protein 3; Uncharacterized protein; cDNA FLJ59124, highly similar to Particularly interesting newCys-His protein; cDNA, FLJ79109, highly similar to Particularly interesting newCys-His protein LIM and senescent cell antigen-like domains 3-like |
| chr1_-_114696472 | 0.08 |
ENST00000393296.1
ENST00000369547.1 ENST00000610222.1 |
SYT6
|
synaptotagmin VI |
| chr9_+_108463234 | 0.08 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr2_-_169887827 | 0.07 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr8_-_125486755 | 0.07 |
ENST00000499418.2
ENST00000530778.1 |
RNF139-AS1
|
RNF139 antisense RNA 1 (head to head) |
| chr6_-_38670897 | 0.07 |
ENST00000373365.4
|
GLO1
|
glyoxalase I |
| chr10_-_96829246 | 0.07 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr13_+_53029564 | 0.07 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr8_+_76452097 | 0.07 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr6_+_108487245 | 0.07 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr2_+_37571717 | 0.07 |
ENST00000338415.3
ENST00000404976.1 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr6_-_52109335 | 0.07 |
ENST00000336123.4
|
IL17F
|
interleukin 17F |
| chr4_-_103746683 | 0.07 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_+_116654376 | 0.07 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr3_+_122785895 | 0.07 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chrX_-_135962876 | 0.07 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chrX_+_149867681 | 0.07 |
ENST00000438018.1
ENST00000436701.1 |
MTMR1
|
myotubularin related protein 1 |
| chr8_+_97597148 | 0.07 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr6_+_12290586 | 0.07 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr4_-_103746924 | 0.07 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr8_+_26150628 | 0.07 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr14_-_92247032 | 0.07 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr16_+_53412368 | 0.07 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr6_-_90348440 | 0.07 |
ENST00000520441.1
ENST00000520318.1 ENST00000523377.1 |
LYRM2
|
LYR motif containing 2 |
| chr12_+_130646999 | 0.07 |
ENST00000539839.1
ENST00000229030.4 |
FZD10
|
frizzled family receptor 10 |
| chr14_+_56584414 | 0.07 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr4_+_169013666 | 0.07 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr1_-_161207986 | 0.07 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr6_+_80714332 | 0.07 |
ENST00000502580.1
ENST00000511260.1 |
TTK
|
TTK protein kinase |
| chr6_-_52859046 | 0.07 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr3_-_112356944 | 0.07 |
ENST00000461431.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr3_-_27498235 | 0.07 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr11_+_112047087 | 0.07 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr4_-_84035905 | 0.07 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr15_-_101817492 | 0.07 |
ENST00000528346.1
ENST00000531964.1 |
VIMP
|
VCP-interacting membrane protein |
| chr3_-_138048682 | 0.07 |
ENST00000383180.2
|
NME9
|
NME/NM23 family member 9 |
| chr5_-_159846066 | 0.07 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr6_-_52859968 | 0.07 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr19_+_41281282 | 0.07 |
ENST00000263369.3
|
MIA
|
melanoma inhibitory activity |
| chr21_+_25801041 | 0.06 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr2_-_69180083 | 0.06 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr1_-_68962805 | 0.06 |
ENST00000370966.5
|
DEPDC1
|
DEP domain containing 1 |
| chr13_+_53030107 | 0.06 |
ENST00000490903.1
ENST00000480747.1 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr19_-_5903714 | 0.06 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr11_+_85566422 | 0.06 |
ENST00000342404.3
|
CCDC83
|
coiled-coil domain containing 83 |
| chr1_+_171107241 | 0.06 |
ENST00000236166.3
|
FMO6P
|
flavin containing monooxygenase 6 pseudogene |
| chrX_-_139866723 | 0.06 |
ENST00000370532.2
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa |
| chr16_-_15472151 | 0.06 |
ENST00000360151.4
ENST00000543801.1 |
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr17_-_62340581 | 0.06 |
ENST00000258991.3
ENST00000583738.1 ENST00000584379.1 |
TEX2
|
testis expressed 2 |
| chr2_+_64068844 | 0.06 |
ENST00000337130.5
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr14_+_56078695 | 0.06 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr8_+_125486939 | 0.06 |
ENST00000303545.3
|
RNF139
|
ring finger protein 139 |
| chr7_+_40174565 | 0.06 |
ENST00000309930.5
ENST00000401647.2 ENST00000335693.4 ENST00000413931.1 ENST00000416370.1 ENST00000540834.1 |
C7orf10
|
succinylCoA:glutarate-CoA transferase |
| chr18_+_19192228 | 0.06 |
ENST00000300413.5
ENST00000579618.1 ENST00000582475.1 |
SNRPD1
|
small nuclear ribonucleoprotein D1 polypeptide 16kDa |
| chr3_-_112127981 | 0.06 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr13_-_31736132 | 0.06 |
ENST00000429785.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr11_-_3400442 | 0.06 |
ENST00000429541.2
ENST00000532539.1 |
ZNF195
|
zinc finger protein 195 |
| chr1_+_92545862 | 0.06 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr10_-_32345305 | 0.06 |
ENST00000302418.4
|
KIF5B
|
kinesin family member 5B |
| chr17_-_27188984 | 0.06 |
ENST00000582320.2
|
MIR144
|
microRNA 451b |
| chr3_+_184534994 | 0.06 |
ENST00000441141.1
ENST00000445089.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr12_-_18243075 | 0.06 |
ENST00000536890.1
|
RERGL
|
RERG/RAS-like |
| chr7_+_35756092 | 0.06 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chr3_+_157154578 | 0.06 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr9_+_130026756 | 0.06 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr12_-_76377795 | 0.06 |
ENST00000552856.1
|
RP11-114H23.1
|
RP11-114H23.1 |
| chr4_+_174089904 | 0.06 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr10_+_21823079 | 0.06 |
ENST00000377100.3
ENST00000377072.3 ENST00000446906.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr12_-_48099773 | 0.06 |
ENST00000432584.3
ENST00000005386.3 |
RPAP3
|
RNA polymerase II associated protein 3 |
| chr14_+_77564440 | 0.06 |
ENST00000361786.2
ENST00000555437.1 ENST00000555611.1 ENST00000554658.1 |
KIAA1737
|
CLOCK-interacting pacemaker |
| chr6_+_153552455 | 0.06 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr11_+_117947724 | 0.06 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr8_-_90996459 | 0.06 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chr10_+_102222798 | 0.06 |
ENST00000343737.5
|
WNT8B
|
wingless-type MMTV integration site family, member 8B |
| chr12_+_16500037 | 0.06 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr10_-_10504285 | 0.06 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr8_-_1922789 | 0.06 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chrX_+_123097014 | 0.05 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr2_-_61245363 | 0.05 |
ENST00000316752.6
|
PUS10
|
pseudouridylate synthase 10 |
| chr11_+_113185778 | 0.05 |
ENST00000524580.2
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr8_+_16884740 | 0.05 |
ENST00000318063.5
|
MICU3
|
mitochondrial calcium uptake family, member 3 |
| chr9_+_103189405 | 0.05 |
ENST00000395067.2
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr11_-_625163 | 0.05 |
ENST00000349570.7
|
CDHR5
|
cadherin-related family member 5 |
| chr9_-_115095123 | 0.05 |
ENST00000458258.1
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr6_-_131299929 | 0.05 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr5_-_175815565 | 0.05 |
ENST00000509257.1
ENST00000507413.1 ENST00000510123.1 |
NOP16
|
NOP16 nucleolar protein |
| chr15_+_36887069 | 0.05 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr14_-_38028689 | 0.05 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr4_-_103998439 | 0.05 |
ENST00000503230.1
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr6_-_8102714 | 0.05 |
ENST00000502429.1
ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr11_+_76571911 | 0.05 |
ENST00000534206.1
ENST00000532485.1 ENST00000526597.1 ENST00000533873.1 ENST00000538157.1 |
ACER3
|
alkaline ceramidase 3 |
| chr7_+_134464376 | 0.05 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr5_+_95997918 | 0.05 |
ENST00000395812.2
ENST00000395813.1 ENST00000359176.4 ENST00000325674.7 |
CAST
|
calpastatin |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD11_HOXA11
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 0.2 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 0.1 | 0.2 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.3 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.2 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.0 | 0.1 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.5 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0033384 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.1 | GO:0045423 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.0 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.0 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.0 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.0 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.0 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.2 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 0.2 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.3 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.0 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.0 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.0 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.0 | GO:0004423 | iduronate-2-sulfatase activity(GO:0004423) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.0 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.0 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.0 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.0 | GO:0031433 | telethonin binding(GO:0031433) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST ADRENERGIC | Adrenergic Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |