|
chr18_+_1099004
Show fit
|
0.89 |
ENST00000581556.1
|
RP11-78F17.1
|
RP11-78F17.1
|
|
chr17_-_46690839
Show fit
|
0.89 |
ENST00000498634.2
|
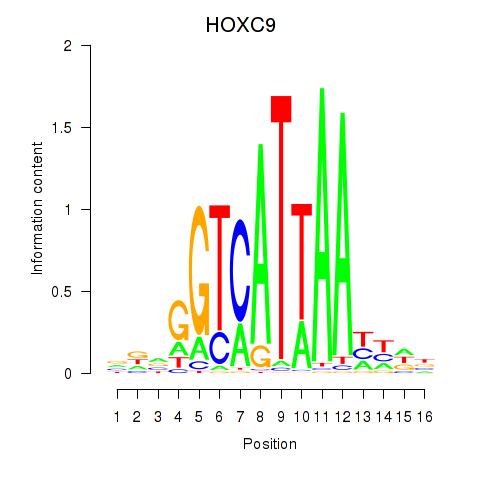
HOXB8
|
homeobox B8
|
|
chr22_-_32767017
Show fit
|
0.79 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense
|
|
chr12_-_91546926
Show fit
|
0.62 |
ENST00000550758.1
|
DCN
|
decorin
|
|
chr9_-_37384431
Show fit
|
0.60 |
ENST00000452923.1
|
RP11-397D12.4
|
RP11-397D12.4
|
|
chr16_+_58074069
Show fit
|
0.57 |
ENST00000570065.1
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted)
|
|
chr2_-_4703793
Show fit
|
0.50 |
ENST00000421212.1
ENST00000412134.1
|
AC022311.1
|
AC022311.1
|
|
chr14_+_39944025
Show fit
|
0.45 |
ENST00000554328.1
ENST00000556620.1
ENST00000557197.1
|
RP11-111A21.1
|
RP11-111A21.1
|
|
chrX_+_13671225
Show fit
|
0.44 |
ENST00000545566.1
ENST00000544987.1
ENST00000314720.4
|
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing
|
|
chr5_-_65018834
Show fit
|
0.43 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta
|
|
chr8_+_52730143
Show fit
|
0.42 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein
|
|
chr20_+_43803517
Show fit
|
0.41 |
ENST00000243924.3
|
PI3
|
peptidase inhibitor 3, skin-derived
|
|
chr12_+_25348139
Show fit
|
0.40 |
ENST00000557540.2
ENST00000381356.4
|
LYRM5
|
LYR motif containing 5
|
|
chr6_-_52859046
Show fit
|
0.39 |
ENST00000457564.1
ENST00000541324.1
ENST00000370960.1
|
GSTA4
|
glutathione S-transferase alpha 4
|
|
chr2_+_37571845
Show fit
|
0.38 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr3_+_122785895
Show fit
|
0.38 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5
|
|
chr13_-_52027134
Show fit
|
0.38 |
ENST00000311234.4
ENST00000425000.1
ENST00000463928.1
ENST00000442263.3
ENST00000398119.2
|
INTS6
|
integrator complex subunit 6
|
|
chr8_-_81083890
Show fit
|
0.38 |
ENST00000518937.1
|
TPD52
|
tumor protein D52
|
|
chr12_+_104235229
Show fit
|
0.36 |
ENST00000551650.1
|
RP11-650K20.3
|
Uncharacterized protein
|
|
chr12_+_25348186
Show fit
|
0.36 |
ENST00000555711.1
ENST00000556885.1
ENST00000554266.1
ENST00000556351.1
ENST00000556927.1
ENST00000556402.1
ENST00000553788.1
|
LYRM5
|
LYR motif containing 5
|
|
chr10_-_13344341
Show fit
|
0.35 |
ENST00000396920.3
|
PHYH
|
phytanoyl-CoA 2-hydroxylase
|
|
chr4_+_56815102
Show fit
|
0.35 |
ENST00000257287.4
|
CEP135
|
centrosomal protein 135kDa
|
|
chr12_+_51347705
Show fit
|
0.34 |
ENST00000398455.3
|
HIGD1C
|
HIG1 hypoxia inducible domain family, member 1C
|
|
chr17_-_38956205
Show fit
|
0.33 |
ENST00000306658.7
|
KRT28
|
keratin 28
|
|
chr8_+_66955648
Show fit
|
0.33 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta
|
|
chr11_+_73498898
Show fit
|
0.33 |
ENST00000535529.1
ENST00000497094.2
ENST00000411840.2
ENST00000535277.1
ENST00000398483.3
ENST00000542303.1
|
MRPL48
|
mitochondrial ribosomal protein L48
|
|
chr2_+_62132781
Show fit
|
0.33 |
ENST00000311832.5
|
COMMD1
|
copper metabolism (Murr1) domain containing 1
|
|
chr11_-_3400442
Show fit
|
0.32 |
ENST00000429541.2
ENST00000532539.1
|
ZNF195
|
zinc finger protein 195
|
|
chr8_-_80993010
Show fit
|
0.32 |
ENST00000537855.1
ENST00000520527.1
ENST00000517427.1
ENST00000448733.2
ENST00000379097.3
|
TPD52
|
tumor protein D52
|
|
chr2_+_109271481
Show fit
|
0.32 |
ENST00000542845.1
ENST00000393314.2
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr2_+_109204909
Show fit
|
0.32 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr16_+_53412368
Show fit
|
0.32 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2
|
|
chr5_+_136070614
Show fit
|
0.32 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1
|
|
chr2_+_38177575
Show fit
|
0.31 |
ENST00000407257.1
ENST00000417700.2
ENST00000234195.3
ENST00000442857.1
|
RMDN2
|
regulator of microtubule dynamics 2
|
|
chr8_+_16884740
Show fit
|
0.31 |
ENST00000318063.5
|
MICU3
|
mitochondrial calcium uptake family, member 3
|
|
chr2_+_162016916
Show fit
|
0.31 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr7_-_156685841
Show fit
|
0.31 |
ENST00000354505.4
ENST00000540390.1
|
LMBR1
|
limb development membrane protein 1
|
|
chr22_-_32766972
Show fit
|
0.30 |
ENST00000382084.4
ENST00000382086.2
|
RFPL3S
|
RFPL3 antisense
|
|
chrX_+_43515467
Show fit
|
0.30 |
ENST00000338702.3
ENST00000542639.1
|
MAOA
|
monoamine oxidase A
|
|
chr5_+_169011033
Show fit
|
0.30 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1
|
|
chr16_-_12897642
Show fit
|
0.29 |
ENST00000433677.2
ENST00000261660.4
ENST00000381774.4
|
CPPED1
|
calcineurin-like phosphoesterase domain containing 1
|
|
chr8_-_93978216
Show fit
|
0.29 |
ENST00000517751.1
ENST00000524107.1
|
TRIQK
|
triple QxxK/R motif containing
|
|
chr9_+_137979506
Show fit
|
0.28 |
ENST00000539529.1
ENST00000392991.4
ENST00000371793.3
|
OLFM1
|
olfactomedin 1
|
|
chr4_+_187812086
Show fit
|
0.28 |
ENST00000507644.2
|
RP11-11N5.1
|
RP11-11N5.1
|
|
chr12_-_21757774
Show fit
|
0.28 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver)
|
|
chr7_+_64838786
Show fit
|
0.28 |
ENST00000450302.2
|
ZNF92
|
zinc finger protein 92
|
|
chr18_-_74839891
Show fit
|
0.28 |
ENST00000581878.1
|
MBP
|
myelin basic protein
|
|
chr8_+_92114060
Show fit
|
0.27 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69
|
|
chr8_-_81083731
Show fit
|
0.27 |
ENST00000379096.5
|
TPD52
|
tumor protein D52
|
|
chr14_+_20187174
Show fit
|
0.27 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2
|
|
chr19_-_23869999
Show fit
|
0.27 |
ENST00000601935.1
ENST00000359788.4
ENST00000600313.1
ENST00000596211.1
ENST00000599168.1
|
ZNF675
|
zinc finger protein 675
|
|
chr7_+_64838712
Show fit
|
0.27 |
ENST00000328747.7
ENST00000431504.1
ENST00000357512.2
|
ZNF92
|
zinc finger protein 92
|
|
chr16_+_28505955
Show fit
|
0.26 |
ENST00000564831.1
ENST00000328423.5
ENST00000431282.1
|
APOBR
|
apolipoprotein B receptor
|
|
chr6_-_52859968
Show fit
|
0.26 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4
|
|
chr11_-_3400330
Show fit
|
0.26 |
ENST00000427810.2
ENST00000005082.9
ENST00000534569.1
ENST00000438262.2
ENST00000528796.1
ENST00000528410.1
ENST00000529678.1
ENST00000354599.6
ENST00000526601.1
ENST00000525502.1
ENST00000533036.1
ENST00000399602.4
|
ZNF195
|
zinc finger protein 195
|
|
chr18_+_32558208
Show fit
|
0.26 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr3_+_56591184
Show fit
|
0.26 |
ENST00000422222.1
ENST00000394672.3
ENST00000326595.7
|
CCDC66
|
coiled-coil domain containing 66
|
|
chr7_-_46736720
Show fit
|
0.25 |
ENST00000451905.1
|
AC011294.3
|
Uncharacterized protein
|
|
chr8_-_93978309
Show fit
|
0.25 |
ENST00000517858.1
ENST00000378861.5
|
TRIQK
|
triple QxxK/R motif containing
|
|
chr8_-_93978346
Show fit
|
0.25 |
ENST00000523580.1
|
TRIQK
|
triple QxxK/R motif containing
|
|
chr7_-_156685890
Show fit
|
0.25 |
ENST00000353442.5
|
LMBR1
|
limb development membrane protein 1
|
|
chr5_+_61874562
Show fit
|
0.25 |
ENST00000334994.5
ENST00000409534.1
|
LRRC70
IPO11
|
leucine rich repeat containing 70
importin 11
|
|
chr12_+_75874580
Show fit
|
0.25 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr16_-_74734742
Show fit
|
0.24 |
ENST00000308807.7
ENST00000573267.1
|
MLKL
|
mixed lineage kinase domain-like
|
|
chr8_-_93978357
Show fit
|
0.24 |
ENST00000522925.1
ENST00000522903.1
ENST00000537541.1
ENST00000518748.1
ENST00000519069.1
ENST00000521988.1
|
TRIQK
|
triple QxxK/R motif containing
|
|
chr2_-_111230393
Show fit
|
0.24 |
ENST00000447537.2
ENST00000413601.2
|
LIMS3L
LIMS3L
|
LIM and senescent cell antigen-like-containing domain protein 3; Uncharacterized protein; cDNA FLJ59124, highly similar to Particularly interesting newCys-His protein; cDNA, FLJ79109, highly similar to Particularly interesting newCys-His protein
LIM and senescent cell antigen-like domains 3-like
|
|
chr7_+_110731062
Show fit
|
0.24 |
ENST00000308478.5
ENST00000451085.1
ENST00000422987.3
ENST00000421101.1
|
LRRN3
|
leucine rich repeat neuronal 3
|
|
chr14_+_62164340
Show fit
|
0.24 |
ENST00000557538.1
ENST00000539097.1
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr8_+_91233744
Show fit
|
0.23 |
ENST00000524361.1
|
LINC00534
|
long intergenic non-protein coding RNA 534
|
|
chr2_+_48541776
Show fit
|
0.23 |
ENST00000413569.1
ENST00000340553.3
|
FOXN2
|
forkhead box N2
|
|
chr3_+_87276407
Show fit
|
0.23 |
ENST00000471660.1
ENST00000263780.4
ENST00000494980.1
|
CHMP2B
|
charged multivesicular body protein 2B
|
|
chr2_+_162016827
Show fit
|
0.23 |
ENST00000429217.1
ENST00000406287.1
ENST00000402568.1
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr2_+_37571717
Show fit
|
0.23 |
ENST00000338415.3
ENST00000404976.1
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr19_+_30433372
Show fit
|
0.23 |
ENST00000312051.6
|
URI1
|
URI1, prefoldin-like chaperone
|
|
chr4_+_56814968
Show fit
|
0.23 |
ENST00000422247.2
|
CEP135
|
centrosomal protein 135kDa
|
|
chr5_-_145562147
Show fit
|
0.23 |
ENST00000545646.1
ENST00000274562.9
ENST00000510191.1
ENST00000394434.2
|
LARS
|
leucyl-tRNA synthetase
|
|
chr8_-_73793975
Show fit
|
0.23 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1
|
|
chr16_+_56782118
Show fit
|
0.23 |
ENST00000566678.1
|
NUP93
|
nucleoporin 93kDa
|
|
chr1_-_54355430
Show fit
|
0.23 |
ENST00000371399.1
ENST00000072644.1
ENST00000412288.1
|
YIPF1
|
Yip1 domain family, member 1
|
|
chr8_-_101718991
Show fit
|
0.22 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1
|
|
chr18_-_32870148
Show fit
|
0.22 |
ENST00000589178.1
ENST00000333206.5
ENST00000592278.1
ENST00000592211.1
ENST00000420878.3
ENST00000383091.2
ENST00000586922.2
|
ZSCAN30
RP11-158H5.7
|
zinc finger and SCAN domain containing 30
RP11-158H5.7
|
|
chr12_+_20963647
Show fit
|
0.22 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr10_+_76871353
Show fit
|
0.22 |
ENST00000542569.1
|
SAMD8
|
sterile alpha motif domain containing 8
|
|
chr5_-_68665296
Show fit
|
0.22 |
ENST00000512152.1
ENST00000503245.1
ENST00000512561.1
ENST00000380822.4
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa
|
|
chr4_-_70626430
Show fit
|
0.22 |
ENST00000310613.3
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1
|
|
chr6_+_83903061
Show fit
|
0.22 |
ENST00000369724.4
ENST00000539997.1
|
RWDD2A
|
RWD domain containing 2A
|
|
chr12_-_8803128
Show fit
|
0.22 |
ENST00000543467.1
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr6_+_76599809
Show fit
|
0.22 |
ENST00000430435.1
|
MYO6
|
myosin VI
|
|
chr4_+_70916119
Show fit
|
0.21 |
ENST00000246896.3
ENST00000511674.1
|
HTN1
|
histatin 1
|
|
chr5_+_176731572
Show fit
|
0.21 |
ENST00000503853.1
|
PRELID1
|
PRELI domain containing 1
|
|
chr5_-_159846066
Show fit
|
0.21 |
ENST00000519349.1
ENST00000520664.1
|
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae)
|
|
chr11_+_73498973
Show fit
|
0.21 |
ENST00000537007.1
|
MRPL48
|
mitochondrial ribosomal protein L48
|
|
chr1_+_35734562
Show fit
|
0.21 |
ENST00000314607.6
ENST00000373297.2
|
ZMYM4
|
zinc finger, MYM-type 4
|
|
chr20_+_57414795
Show fit
|
0.21 |
ENST00000371098.2
ENST00000371075.3
|
GNAS
|
GNAS complex locus
|
|
chr2_-_230096756
Show fit
|
0.21 |
ENST00000354069.6
|
PID1
|
phosphotyrosine interaction domain containing 1
|
|
chr13_-_24007815
Show fit
|
0.21 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr1_+_200993071
Show fit
|
0.20 |
ENST00000446333.1
ENST00000458003.1
|
RP11-168O16.1
|
RP11-168O16.1
|
|
chr9_-_104198042
Show fit
|
0.20 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate
|
|
chr22_+_21133469
Show fit
|
0.20 |
ENST00000406799.1
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1
|
|
chr2_-_8723918
Show fit
|
0.20 |
ENST00000454224.1
|
AC011747.4
|
AC011747.4
|
|
chr3_-_25824872
Show fit
|
0.20 |
ENST00000308710.5
|
NGLY1
|
N-glycanase 1
|
|
chr8_-_93978333
Show fit
|
0.20 |
ENST00000524037.1
ENST00000520430.1
ENST00000521617.1
|
TRIQK
|
triple QxxK/R motif containing
|
|
chr9_+_70856899
Show fit
|
0.20 |
ENST00000377342.5
ENST00000478048.1
|
CBWD3
|
COBW domain containing 3
|
|
chr6_+_131571535
Show fit
|
0.20 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr10_+_76871229
Show fit
|
0.20 |
ENST00000372690.3
|
SAMD8
|
sterile alpha motif domain containing 8
|
|
chr3_-_124653579
Show fit
|
0.19 |
ENST00000478191.1
ENST00000311075.3
|
MUC13
|
mucin 13, cell surface associated
|
|
chr20_+_57414743
Show fit
|
0.19 |
ENST00000313949.7
|
GNAS
|
GNAS complex locus
|
|
chr2_-_17981462
Show fit
|
0.19 |
ENST00000402989.1
ENST00000428868.1
|
SMC6
|
structural maintenance of chromosomes 6
|
|
chr19_-_48753028
Show fit
|
0.19 |
ENST00000522431.1
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr4_+_71600144
Show fit
|
0.19 |
ENST00000502653.1
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr7_-_44580861
Show fit
|
0.19 |
ENST00000546276.1
ENST00000289547.4
ENST00000381160.3
ENST00000423141.1
|
NPC1L1
|
NPC1-like 1
|
|
chr4_+_71600063
Show fit
|
0.19 |
ENST00000513597.1
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr5_-_176433693
Show fit
|
0.19 |
ENST00000507513.1
ENST00000511320.1
|
UIMC1
|
ubiquitin interaction motif containing 1
|
|
chr12_-_118628315
Show fit
|
0.19 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3
|
|
chr2_+_109237717
Show fit
|
0.19 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr14_-_92247032
Show fit
|
0.19 |
ENST00000556661.1
ENST00000553676.1
ENST00000554560.1
|
CATSPERB
|
catsper channel auxiliary subunit beta
|
|
chr2_+_162016804
Show fit
|
0.19 |
ENST00000392749.2
ENST00000440506.1
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr1_+_47603109
Show fit
|
0.19 |
ENST00000371890.3
ENST00000294337.3
ENST00000371891.3
|
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 22
|
|
chr2_-_169887827
Show fit
|
0.18 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11
|
|
chr12_+_123259063
Show fit
|
0.18 |
ENST00000392441.4
ENST00000539171.1
|
CCDC62
|
coiled-coil domain containing 62
|
|
chr14_+_55494323
Show fit
|
0.18 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4
|
|
chr15_+_45003675
Show fit
|
0.18 |
ENST00000558401.1
ENST00000559916.1
ENST00000544417.1
|
B2M
|
beta-2-microglobulin
|
|
chr18_-_46895066
Show fit
|
0.18 |
ENST00000583225.1
ENST00000584983.1
ENST00000583280.1
ENST00000581738.1
|
DYM
|
dymeclin
|
|
chr11_+_120039685
Show fit
|
0.18 |
ENST00000530303.1
ENST00000319763.1
|
AP000679.2
|
Uncharacterized protein
|
|
chr6_+_111303218
Show fit
|
0.18 |
ENST00000441448.2
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae)
|
|
chr5_-_148442584
Show fit
|
0.18 |
ENST00000394358.2
ENST00000512049.1
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr1_-_113615699
Show fit
|
0.18 |
ENST00000421157.1
|
RP11-31F15.2
|
RP11-31F15.2
|
|
chr14_+_78174414
Show fit
|
0.18 |
ENST00000557342.1
ENST00000238688.5
ENST00000557623.1
ENST00000557431.1
ENST00000556831.1
ENST00000556375.1
ENST00000553981.1
|
SLIRP
|
SRA stem-loop interacting RNA binding protein
|
|
chr22_+_45714361
Show fit
|
0.18 |
ENST00000452238.1
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr3_-_170587815
Show fit
|
0.17 |
ENST00000466674.1
|
RPL22L1
|
ribosomal protein L22-like 1
|
|
chr1_-_26231589
Show fit
|
0.17 |
ENST00000374291.1
|
STMN1
|
stathmin 1
|
|
chr19_+_21265028
Show fit
|
0.17 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714
|
|
chr10_-_123687431
Show fit
|
0.17 |
ENST00000423243.1
|
ATE1
|
arginyltransferase 1
|
|
chr16_-_74734672
Show fit
|
0.17 |
ENST00000306247.7
ENST00000575686.1
|
MLKL
|
mixed lineage kinase domain-like
|
|
chr2_-_86850949
Show fit
|
0.17 |
ENST00000237455.4
|
RNF103
|
ring finger protein 103
|
|
chr2_+_114195268
Show fit
|
0.17 |
ENST00000259199.4
ENST00000416503.2
ENST00000433343.2
|
CBWD2
|
COBW domain containing 2
|
|
chr11_-_107729887
Show fit
|
0.17 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2
|
|
chr10_+_118608998
Show fit
|
0.17 |
ENST00000409522.1
ENST00000341276.5
ENST00000512864.2
|
ENO4
|
enolase family member 4
|
|
chr2_-_69664586
Show fit
|
0.17 |
ENST00000303698.3
ENST00000394305.1
ENST00000410022.2
|
NFU1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae)
|
|
chr8_-_17579726
Show fit
|
0.17 |
ENST00000381861.3
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr15_-_101817492
Show fit
|
0.17 |
ENST00000528346.1
ENST00000531964.1
|
VIMP
|
VCP-interacting membrane protein
|
|
chr20_-_45980621
Show fit
|
0.17 |
ENST00000446894.1
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr5_-_125930929
Show fit
|
0.16 |
ENST00000553117.1
ENST00000447989.2
ENST00000409134.3
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr2_+_70121075
Show fit
|
0.16 |
ENST00000409116.1
|
SNRNP27
|
small nuclear ribonucleoprotein 27kDa (U4/U6.U5)
|
|
chr4_+_155484103
Show fit
|
0.16 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain
|
|
chr10_+_45495898
Show fit
|
0.16 |
ENST00000298299.3
|
ZNF22
|
zinc finger protein 22
|
|
chr1_-_212965104
Show fit
|
0.16 |
ENST00000422588.2
ENST00000366975.6
ENST00000366977.3
ENST00000366976.1
|
NSL1
|
NSL1, MIS12 kinetochore complex component
|
|
chr7_-_76955563
Show fit
|
0.16 |
ENST00000441833.2
|
GSAP
|
gamma-secretase activating protein
|
|
chr10_+_74927875
Show fit
|
0.15 |
ENST00000242505.6
|
FAM149B1
|
family with sequence similarity 149, member B1
|
|
chr5_-_68665084
Show fit
|
0.15 |
ENST00000509462.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa
|
|
chr2_+_109204743
Show fit
|
0.15 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr3_-_170587974
Show fit
|
0.15 |
ENST00000463836.1
|
RPL22L1
|
ribosomal protein L22-like 1
|
|
chr5_-_35991535
Show fit
|
0.15 |
ENST00000507113.1
ENST00000274278.3
|
UGT3A1
|
UDP glycosyltransferase 3 family, polypeptide A1
|
|
chr5_-_68664989
Show fit
|
0.15 |
ENST00000508954.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa
|
|
chr4_+_86525299
Show fit
|
0.15 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr15_+_52155001
Show fit
|
0.15 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous)
|
|
chr2_+_160590469
Show fit
|
0.15 |
ENST00000409591.1
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase
|
|
chr9_-_179018
Show fit
|
0.15 |
ENST00000431099.2
ENST00000382447.4
ENST00000382389.1
ENST00000377447.3
ENST00000314367.10
ENST00000356521.4
ENST00000382393.1
ENST00000377400.4
|
CBWD1
|
COBW domain containing 1
|
|
chr8_-_17555164
Show fit
|
0.15 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr8_-_62602327
Show fit
|
0.15 |
ENST00000445642.3
ENST00000517847.2
ENST00000389204.4
ENST00000517661.1
ENST00000517903.1
ENST00000522603.1
ENST00000522349.1
ENST00000522835.1
ENST00000541428.1
ENST00000518306.1
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr3_+_143690640
Show fit
|
0.15 |
ENST00000315691.3
|
C3orf58
|
chromosome 3 open reading frame 58
|
|
chr9_-_69262509
Show fit
|
0.15 |
ENST00000377449.1
ENST00000382399.4
ENST00000377439.1
ENST00000377441.1
ENST00000377457.5
|
CBWD6
|
COBW domain containing 6
|
|
chr8_-_30670384
Show fit
|
0.15 |
ENST00000221138.4
ENST00000518243.1
|
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme
|
|
chr1_+_151735431
Show fit
|
0.14 |
ENST00000321531.5
ENST00000315067.8
|
OAZ3
|
ornithine decarboxylase antizyme 3
|
|
chr17_+_58018269
Show fit
|
0.14 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein
|
|
chr6_-_167797887
Show fit
|
0.14 |
ENST00000476779.2
ENST00000460930.2
ENST00000397829.4
ENST00000366827.2
|
TCP10
|
t-complex 10
|
|
chr20_-_48782639
Show fit
|
0.14 |
ENST00000435301.2
|
RP11-112L6.3
|
RP11-112L6.3
|
|
chr8_+_104033277
Show fit
|
0.14 |
ENST00000518857.1
ENST00000395862.3
|
ATP6V1C1
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1
|
|
chr14_-_55907334
Show fit
|
0.13 |
ENST00000247219.5
|
TBPL2
|
TATA box binding protein like 2
|
|
chr7_+_7606497
Show fit
|
0.13 |
ENST00000340080.4
ENST00000405785.1
ENST00000433635.1
|
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila)
|
|
chr10_+_98592009
Show fit
|
0.13 |
ENST00000540664.1
ENST00000371103.3
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr2_-_242212227
Show fit
|
0.13 |
ENST00000427007.1
ENST00000458564.1
ENST00000452065.1
ENST00000427183.2
ENST00000426343.1
ENST00000422080.1
ENST00000449504.1
ENST00000449864.1
ENST00000391975.1
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr10_-_96829246
Show fit
|
0.13 |
ENST00000371270.3
ENST00000535898.1
ENST00000539050.1
|
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8
|
|
chr10_-_31146615
Show fit
|
0.13 |
ENST00000444692.2
|
ZNF438
|
zinc finger protein 438
|
|
chr12_-_104234966
Show fit
|
0.13 |
ENST00000392876.3
|
NT5DC3
|
5'-nucleotidase domain containing 3
|
|
chr16_+_72090053
Show fit
|
0.13 |
ENST00000576168.2
ENST00000567185.3
ENST00000567612.2
|
HP
|
haptoglobin
|
|
chr4_-_103748880
Show fit
|
0.12 |
ENST00000453744.2
ENST00000349311.8
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3
|
|
chr12_+_32687221
Show fit
|
0.12 |
ENST00000525053.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr8_+_76452097
Show fit
|
0.12 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr1_+_40506392
Show fit
|
0.12 |
ENST00000414893.1
ENST00000414281.1
ENST00000420216.1
ENST00000372792.2
ENST00000372798.1
ENST00000340450.3
ENST00000372805.3
ENST00000435719.1
ENST00000427843.1
ENST00000417287.1
ENST00000424977.1
ENST00000446031.1
|
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast)
|
|
chr4_+_71248795
Show fit
|
0.12 |
ENST00000304915.3
|
SMR3B
|
submaxillary gland androgen regulated protein 3B
|
|
chr17_+_28268623
Show fit
|
0.12 |
ENST00000394835.3
ENST00000320856.5
ENST00000394832.2
ENST00000378738.3
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr10_-_94257512
Show fit
|
0.12 |
ENST00000371581.5
|
IDE
|
insulin-degrading enzyme
|
|
chr12_-_30887948
Show fit
|
0.12 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2
|
|
chr18_-_12656715
Show fit
|
0.12 |
ENST00000462226.1
ENST00000497844.2
ENST00000309836.5
ENST00000453447.2
|
SPIRE1
|
spire-type actin nucleation factor 1
|
|
chr4_+_155484155
Show fit
|
0.12 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain
|
|
chr4_+_70894130
Show fit
|
0.12 |
ENST00000526767.1
ENST00000530128.1
ENST00000381057.3
|
HTN3
|
histatin 3
|
|
chr10_-_123687497
Show fit
|
0.12 |
ENST00000369040.3
ENST00000224652.6
ENST00000369043.3
|
ATE1
|
arginyltransferase 1
|
|
chr2_-_128284020
Show fit
|
0.12 |
ENST00000295321.4
ENST00000455721.2
|
IWS1
|
IWS1 homolog (S. cerevisiae)
|
|
chr4_-_103749105
Show fit
|
0.11 |
ENST00000394801.4
ENST00000394804.2
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3
|
|
chr12_-_118628350
Show fit
|
0.11 |
ENST00000537952.1
ENST00000537822.1
|
TAOK3
|
TAO kinase 3
|
|
chr13_+_33160553
Show fit
|
0.11 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae)
|
|
chr14_+_100240019
Show fit
|
0.11 |
ENST00000556199.1
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr9_-_95056010
Show fit
|
0.11 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr1_+_63989004
Show fit
|
0.11 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7
|
|
chr4_-_69215467
Show fit
|
0.11 |
ENST00000579690.1
|
YTHDC1
|
YTH domain containing 1
|
|
chr4_-_52883786
Show fit
|
0.11 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66
|
|
chr14_-_21492113
Show fit
|
0.11 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2
|
|
chr10_-_123687399
Show fit
|
0.11 |
ENST00000543447.1
|
ATE1
|
arginyltransferase 1
|
|
chr15_+_69854027
Show fit
|
0.11 |
ENST00000498938.2
|
RP11-279F6.1
|
RP11-279F6.1
|