Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
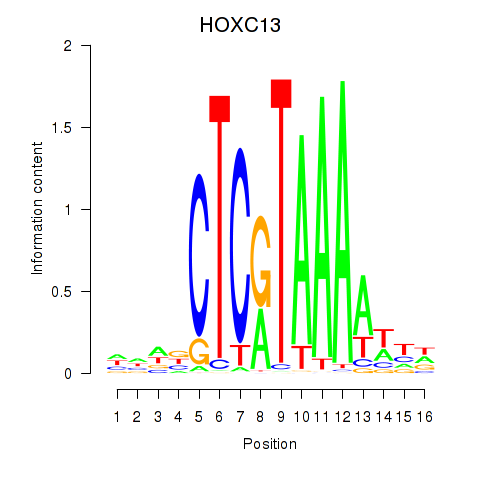
Results for HOXC13
Z-value: 0.83
Transcription factors associated with HOXC13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC13
|
ENSG00000123364.3 | homeobox C13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC13 | hg19_v2_chr12_+_54332535_54332636 | -0.87 | 1.3e-01 | Click! |
Activity profile of HOXC13 motif
Sorted Z-values of HOXC13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_15496722 | 0.63 |
ENST00000472534.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr15_-_71184724 | 0.52 |
ENST00000560604.1
|
THAP10
|
THAP domain containing 10 |
| chr14_-_88200641 | 0.36 |
ENST00000556168.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chr6_+_126221034 | 0.34 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr7_-_64023441 | 0.30 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr14_-_82089405 | 0.30 |
ENST00000554211.1
|
RP11-799P8.1
|
RP11-799P8.1 |
| chr15_+_85523671 | 0.30 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chr15_+_77713299 | 0.30 |
ENST00000559099.1
|
HMG20A
|
high mobility group 20A |
| chr21_+_25801041 | 0.29 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr15_+_71228826 | 0.26 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr3_-_112127981 | 0.25 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr16_-_46865286 | 0.25 |
ENST00000285697.4
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr4_-_142253761 | 0.24 |
ENST00000511213.1
|
RP11-362F19.1
|
RP11-362F19.1 |
| chr9_-_5304432 | 0.23 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chrX_-_38186775 | 0.23 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr3_-_172241250 | 0.23 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr17_-_8286484 | 0.23 |
ENST00000582556.1
ENST00000584164.1 ENST00000293842.5 ENST00000584343.1 ENST00000578812.1 ENST00000583011.1 |
RPL26
|
ribosomal protein L26 |
| chrX_-_100129320 | 0.23 |
ENST00000372966.3
|
NOX1
|
NADPH oxidase 1 |
| chr1_-_154150651 | 0.22 |
ENST00000302206.5
|
TPM3
|
tropomyosin 3 |
| chr19_-_23869999 | 0.22 |
ENST00000601935.1
ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr15_+_77224045 | 0.21 |
ENST00000320963.5
ENST00000394883.3 |
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr1_-_150208498 | 0.21 |
ENST00000314136.8
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr15_+_79166065 | 0.21 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr21_+_47706537 | 0.20 |
ENST00000397691.1
|
YBEY
|
ybeY metallopeptidase (putative) |
| chr3_+_113667354 | 0.20 |
ENST00000491556.1
|
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr7_+_64838712 | 0.20 |
ENST00000328747.7
ENST00000431504.1 ENST00000357512.2 |
ZNF92
|
zinc finger protein 92 |
| chr17_-_56769382 | 0.20 |
ENST00000240361.8
ENST00000349033.5 ENST00000389934.3 |
TEX14
|
testis expressed 14 |
| chr3_-_179169181 | 0.20 |
ENST00000497513.1
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr7_+_64838786 | 0.19 |
ENST00000450302.2
|
ZNF92
|
zinc finger protein 92 |
| chr12_-_13248598 | 0.19 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr18_-_74839891 | 0.19 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr3_-_195310802 | 0.19 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr7_-_25219897 | 0.19 |
ENST00000283905.3
ENST00000409280.1 ENST00000415598.1 |
C7orf31
|
chromosome 7 open reading frame 31 |
| chr5_-_19142450 | 0.19 |
ENST00000504827.1
|
RP11-124N3.3
|
RP11-124N3.3 |
| chr2_+_201390843 | 0.18 |
ENST00000357799.4
ENST00000409203.3 |
SGOL2
|
shugoshin-like 2 (S. pombe) |
| chr1_-_17231271 | 0.18 |
ENST00000606899.1
|
RP11-108M9.6
|
RP11-108M9.6 |
| chr5_+_138678131 | 0.18 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr7_-_64023410 | 0.18 |
ENST00000447137.2
|
ZNF680
|
zinc finger protein 680 |
| chr15_+_71184931 | 0.18 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr14_-_50319758 | 0.17 |
ENST00000298310.5
|
NEMF
|
nuclear export mediator factor |
| chr14_-_50319482 | 0.17 |
ENST00000546046.1
ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF
|
nuclear export mediator factor |
| chr2_+_162016827 | 0.17 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr4_+_128702969 | 0.17 |
ENST00000508776.1
ENST00000439123.2 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr19_-_38085633 | 0.16 |
ENST00000593133.1
ENST00000590751.1 ENST00000358744.3 ENST00000328550.2 ENST00000451802.2 |
ZNF571
|
zinc finger protein 571 |
| chr3_-_160116995 | 0.16 |
ENST00000465537.1
ENST00000486856.1 ENST00000468218.1 ENST00000478370.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr16_+_82068585 | 0.16 |
ENST00000563491.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr2_-_240985489 | 0.16 |
ENST00000319423.4
|
OR6B3
|
olfactory receptor, family 6, subfamily B, member 3 |
| chr4_+_124571409 | 0.16 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr10_+_30723533 | 0.16 |
ENST00000413724.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr4_+_119200215 | 0.16 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr1_+_171107241 | 0.16 |
ENST00000236166.3
|
FMO6P
|
flavin containing monooxygenase 6 pseudogene |
| chr9_+_40028620 | 0.16 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr20_-_52687059 | 0.16 |
ENST00000371435.2
ENST00000395961.3 |
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr4_+_106631966 | 0.15 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr9_+_137979506 | 0.15 |
ENST00000539529.1
ENST00000392991.4 ENST00000371793.3 |
OLFM1
|
olfactomedin 1 |
| chr12_-_123728548 | 0.15 |
ENST00000545406.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr3_+_159557637 | 0.15 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr8_+_101170563 | 0.15 |
ENST00000520508.1
ENST00000388798.2 |
SPAG1
|
sperm associated antigen 1 |
| chr10_-_13544945 | 0.15 |
ENST00000378605.3
ENST00000341083.3 |
BEND7
|
BEN domain containing 7 |
| chr17_+_39382900 | 0.15 |
ENST00000377721.3
ENST00000455970.2 |
KRTAP9-2
|
keratin associated protein 9-2 |
| chr19_+_21265028 | 0.15 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714 |
| chr7_-_69062391 | 0.15 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr19_-_6279932 | 0.15 |
ENST00000252674.7
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
| chr7_-_27169801 | 0.14 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr4_-_17513604 | 0.14 |
ENST00000505710.1
|
QDPR
|
quinoid dihydropteridine reductase |
| chr12_-_91546926 | 0.14 |
ENST00000550758.1
|
DCN
|
decorin |
| chr1_+_219347203 | 0.14 |
ENST00000366927.3
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr6_+_168434678 | 0.14 |
ENST00000496008.1
|
KIF25
|
kinesin family member 25 |
| chr2_+_162016916 | 0.14 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr15_+_71185148 | 0.14 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr19_+_11350278 | 0.14 |
ENST00000252453.8
|
C19orf80
|
chromosome 19 open reading frame 80 |
| chr6_-_75953484 | 0.14 |
ENST00000472311.2
ENST00000460985.1 ENST00000377978.3 ENST00000509698.1 ENST00000230459.4 ENST00000370089.2 |
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr3_-_98241760 | 0.14 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr4_+_54243917 | 0.14 |
ENST00000507166.1
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr2_+_48010312 | 0.14 |
ENST00000540021.1
|
MSH6
|
mutS homolog 6 |
| chr10_+_13628933 | 0.13 |
ENST00000417658.1
ENST00000320054.4 |
PRPF18
|
pre-mRNA processing factor 18 |
| chr1_+_222886694 | 0.13 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr11_+_85566422 | 0.13 |
ENST00000342404.3
|
CCDC83
|
coiled-coil domain containing 83 |
| chr12_-_4647606 | 0.13 |
ENST00000261250.3
ENST00000541014.1 ENST00000545746.1 ENST00000542080.1 |
C12orf4
|
chromosome 12 open reading frame 4 |
| chr17_+_56769924 | 0.13 |
ENST00000461271.1
ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C
|
RAD51 paralog C |
| chr10_-_31288398 | 0.13 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr6_-_112080256 | 0.12 |
ENST00000462856.2
ENST00000229471.4 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chrX_+_13707235 | 0.12 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr19_+_45409011 | 0.12 |
ENST00000252486.4
ENST00000446996.1 ENST00000434152.1 |
APOE
|
apolipoprotein E |
| chr5_+_134181625 | 0.12 |
ENST00000394976.3
|
C5orf24
|
chromosome 5 open reading frame 24 |
| chrM_+_8527 | 0.12 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr5_+_101569696 | 0.12 |
ENST00000597120.1
|
AC008948.1
|
AC008948.1 |
| chr1_+_174844645 | 0.12 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr4_+_8321882 | 0.12 |
ENST00000509453.1
ENST00000503186.1 |
RP11-774O3.2
RP11-774O3.1
|
RP11-774O3.2 RP11-774O3.1 |
| chr5_+_72509751 | 0.12 |
ENST00000515556.1
ENST00000513379.1 ENST00000427584.2 |
RP11-60A8.1
|
RP11-60A8.1 |
| chr2_+_61372226 | 0.12 |
ENST00000426997.1
|
C2orf74
|
chromosome 2 open reading frame 74 |
| chr1_+_46769303 | 0.12 |
ENST00000311672.5
|
UQCRH
|
ubiquinol-cytochrome c reductase hinge protein |
| chr3_+_99986036 | 0.12 |
ENST00000471098.1
|
TBC1D23
|
TBC1 domain family, member 23 |
| chr6_-_32784687 | 0.11 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr2_+_172778952 | 0.11 |
ENST00000392584.1
ENST00000264108.4 |
HAT1
|
histone acetyltransferase 1 |
| chr9_-_2844058 | 0.11 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr11_+_101785727 | 0.11 |
ENST00000263468.8
|
KIAA1377
|
KIAA1377 |
| chr3_+_100428188 | 0.11 |
ENST00000418917.2
ENST00000490574.1 |
TFG
|
TRK-fused gene |
| chr9_+_112887772 | 0.11 |
ENST00000259318.7
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr17_-_39526052 | 0.11 |
ENST00000251646.3
|
KRT33B
|
keratin 33B |
| chr16_-_72206034 | 0.11 |
ENST00000537465.1
ENST00000237353.10 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr5_+_169010638 | 0.11 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr4_-_170533723 | 0.11 |
ENST00000510533.1
ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1
|
NIMA-related kinase 1 |
| chr22_-_18111499 | 0.10 |
ENST00000413576.1
ENST00000399796.2 ENST00000399798.2 ENST00000253413.5 |
ATP6V1E1
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E1 |
| chr2_+_162016804 | 0.10 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr3_+_100428316 | 0.10 |
ENST00000479672.1
ENST00000476228.1 ENST00000463568.1 |
TFG
|
TRK-fused gene |
| chr17_+_43861680 | 0.10 |
ENST00000314537.5
|
CRHR1
|
corticotropin releasing hormone receptor 1 |
| chr1_-_150208412 | 0.10 |
ENST00000532744.1
ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr6_-_33239712 | 0.10 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr1_+_206858232 | 0.10 |
ENST00000294981.4
|
MAPKAPK2
|
mitogen-activated protein kinase-activated protein kinase 2 |
| chr6_-_130031358 | 0.10 |
ENST00000368149.2
|
ARHGAP18
|
Rho GTPase activating protein 18 |
| chr7_-_32534850 | 0.10 |
ENST00000409952.3
ENST00000409909.3 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr4_-_184241927 | 0.10 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr14_+_73634537 | 0.10 |
ENST00000406768.1
|
PSEN1
|
presenilin 1 |
| chr18_-_46895066 | 0.10 |
ENST00000583225.1
ENST00000584983.1 ENST00000583280.1 ENST00000581738.1 |
DYM
|
dymeclin |
| chrX_+_41583408 | 0.10 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr5_-_180076613 | 0.10 |
ENST00000261937.6
ENST00000393347.3 |
FLT4
|
fms-related tyrosine kinase 4 |
| chr3_-_51533966 | 0.09 |
ENST00000504652.1
|
VPRBP
|
Vpr (HIV-1) binding protein |
| chr16_+_28565230 | 0.09 |
ENST00000317058.3
|
CCDC101
|
coiled-coil domain containing 101 |
| chr1_+_47603109 | 0.09 |
ENST00000371890.3
ENST00000294337.3 ENST00000371891.3 |
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| chrY_+_14958970 | 0.09 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr7_-_152373216 | 0.09 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr6_+_88182643 | 0.09 |
ENST00000369556.3
ENST00000544441.1 ENST00000369552.4 ENST00000369557.5 |
SLC35A1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1 |
| chr6_+_56820152 | 0.09 |
ENST00000370745.1
|
BEND6
|
BEN domain containing 6 |
| chr22_+_32439019 | 0.09 |
ENST00000266088.4
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr11_-_47546220 | 0.09 |
ENST00000528538.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr11_+_94706804 | 0.09 |
ENST00000335080.5
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr5_-_79287060 | 0.09 |
ENST00000512560.1
ENST00000509852.1 ENST00000512528.1 |
MTX3
|
metaxin 3 |
| chr12_+_45609893 | 0.09 |
ENST00000320560.8
|
ANO6
|
anoctamin 6 |
| chr15_+_77223960 | 0.09 |
ENST00000394885.3
|
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr1_-_168464875 | 0.09 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr13_+_34392185 | 0.09 |
ENST00000380071.3
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr14_-_92333873 | 0.09 |
ENST00000435962.2
|
TC2N
|
tandem C2 domains, nuclear |
| chr10_-_79398250 | 0.09 |
ENST00000286627.5
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr5_+_56471592 | 0.08 |
ENST00000511209.1
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr9_-_88714421 | 0.08 |
ENST00000388712.3
|
GOLM1
|
golgi membrane protein 1 |
| chr13_+_103451399 | 0.08 |
ENST00000257336.1
ENST00000448849.2 |
BIVM
|
basic, immunoglobulin-like variable motif containing |
| chr20_-_52687030 | 0.08 |
ENST00000411563.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr5_-_39270725 | 0.08 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr9_-_88715044 | 0.08 |
ENST00000388711.3
ENST00000466178.1 |
GOLM1
|
golgi membrane protein 1 |
| chr12_+_45609862 | 0.08 |
ENST00000423947.3
ENST00000435642.1 |
ANO6
|
anoctamin 6 |
| chr10_+_70847852 | 0.08 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr1_-_150208363 | 0.08 |
ENST00000436748.2
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr2_+_33359687 | 0.08 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr2_-_37384175 | 0.08 |
ENST00000411537.2
ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr4_+_141178440 | 0.08 |
ENST00000394205.3
|
SCOC
|
short coiled-coil protein |
| chr1_-_55266926 | 0.08 |
ENST00000371276.4
|
TTC22
|
tetratricopeptide repeat domain 22 |
| chr9_+_123884038 | 0.07 |
ENST00000373847.1
|
CNTRL
|
centriolin |
| chr12_-_53207842 | 0.07 |
ENST00000458244.2
|
KRT4
|
keratin 4 |
| chr19_+_52102540 | 0.07 |
ENST00000601315.1
|
AC018755.17
|
AC018755.17 |
| chr10_+_94352956 | 0.07 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr2_-_165424973 | 0.07 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr14_-_24701539 | 0.07 |
ENST00000534348.1
ENST00000524927.1 ENST00000250495.5 |
NEDD8-MDP1
NEDD8
|
NEDD8-MDP1 readthrough neural precursor cell expressed, developmentally down-regulated 8 |
| chr10_-_113943447 | 0.07 |
ENST00000369425.1
ENST00000348367.4 ENST00000423155.1 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr2_-_100925967 | 0.07 |
ENST00000409647.1
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr19_+_16999654 | 0.07 |
ENST00000248076.3
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr2_+_33359646 | 0.07 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr2_-_223520770 | 0.07 |
ENST00000536361.1
|
FARSB
|
phenylalanyl-tRNA synthetase, beta subunit |
| chr13_-_103451307 | 0.07 |
ENST00000376004.4
|
KDELC1
|
KDEL (Lys-Asp-Glu-Leu) containing 1 |
| chr10_-_127464390 | 0.07 |
ENST00000368808.3
|
MMP21
|
matrix metallopeptidase 21 |
| chr3_+_57875738 | 0.07 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chr20_+_19870167 | 0.07 |
ENST00000440354.2
|
RIN2
|
Ras and Rab interactor 2 |
| chr9_-_32526299 | 0.07 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr6_+_127898312 | 0.07 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr6_+_21666633 | 0.07 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr4_+_122722466 | 0.06 |
ENST00000243498.5
ENST00000379663.3 ENST00000509800.1 |
EXOSC9
|
exosome component 9 |
| chr7_+_23749854 | 0.06 |
ENST00000456014.2
|
STK31
|
serine/threonine kinase 31 |
| chr1_-_220220000 | 0.06 |
ENST00000366923.3
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr1_-_86174065 | 0.06 |
ENST00000370574.3
ENST00000431532.2 |
ZNHIT6
|
zinc finger, HIT-type containing 6 |
| chr1_-_13331692 | 0.06 |
ENST00000353410.5
ENST00000376173.3 |
PRAMEF3
|
PRAME family member 3 |
| chr19_-_57656570 | 0.06 |
ENST00000269834.1
|
ZIM3
|
zinc finger, imprinted 3 |
| chr13_+_34392200 | 0.06 |
ENST00000434425.1
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr4_+_70894130 | 0.06 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr17_-_76824975 | 0.06 |
ENST00000586066.2
|
USP36
|
ubiquitin specific peptidase 36 |
| chr9_-_130533615 | 0.06 |
ENST00000373277.4
|
SH2D3C
|
SH2 domain containing 3C |
| chr22_+_18111594 | 0.06 |
ENST00000399782.1
|
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr2_+_33359473 | 0.06 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr17_-_28257080 | 0.06 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr2_+_138721850 | 0.06 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr6_-_128222103 | 0.06 |
ENST00000434358.1
ENST00000543064.1 ENST00000368248.2 |
THEMIS
|
thymocyte selection associated |
| chr3_-_71179699 | 0.06 |
ENST00000497355.1
|
FOXP1
|
forkhead box P1 |
| chr14_-_80678512 | 0.06 |
ENST00000553968.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr2_+_33683109 | 0.06 |
ENST00000437184.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr11_+_107643129 | 0.06 |
ENST00000447610.1
|
AP001024.2
|
Uncharacterized protein |
| chr12_+_75874580 | 0.06 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr7_-_30066233 | 0.06 |
ENST00000222803.5
|
FKBP14
|
FK506 binding protein 14, 22 kDa |
| chr2_-_86850949 | 0.06 |
ENST00000237455.4
|
RNF103
|
ring finger protein 103 |
| chr5_-_58571935 | 0.05 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr9_-_69229650 | 0.05 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr4_+_175204818 | 0.05 |
ENST00000503780.1
|
CEP44
|
centrosomal protein 44kDa |
| chr17_+_35732955 | 0.05 |
ENST00000300618.4
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr3_-_160117035 | 0.05 |
ENST00000489004.1
ENST00000496589.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr11_+_7618413 | 0.05 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr5_-_180076580 | 0.05 |
ENST00000502649.1
|
FLT4
|
fms-related tyrosine kinase 4 |
| chr1_+_90460661 | 0.05 |
ENST00000340281.4
ENST00000361911.5 ENST00000370447.3 ENST00000455342.2 |
ZNF326
|
zinc finger protein 326 |
| chr7_+_23749822 | 0.05 |
ENST00000422637.1
|
STK31
|
serine/threonine kinase 31 |
| chr14_+_23727694 | 0.05 |
ENST00000399905.1
ENST00000470456.1 |
C14orf164
|
chromosome 14 open reading frame 164 |
| chr7_-_76255444 | 0.05 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr1_+_174128639 | 0.05 |
ENST00000251507.4
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr10_-_6019984 | 0.05 |
ENST00000525219.2
|
IL15RA
|
interleukin 15 receptor, alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.2 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.1 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.2 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.2 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.1 | GO:2000646 | lipid transport involved in lipid storage(GO:0010877) regulation of Cdc42 protein signal transduction(GO:0032489) positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.2 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:1902231 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.2 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.1 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.1 | GO:1904796 | regulation of core promoter binding(GO:1904796) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.2 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.0 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:2000849 | glucocorticoid secretion(GO:0035933) regulation of glucocorticoid secretion(GO:2000849) |
| 0.0 | 0.0 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.0 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.0 | 0.1 | GO:0036328 | VEGF-C-activated receptor activity(GO:0036328) |
| 0.0 | 0.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.1 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.0 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |