Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
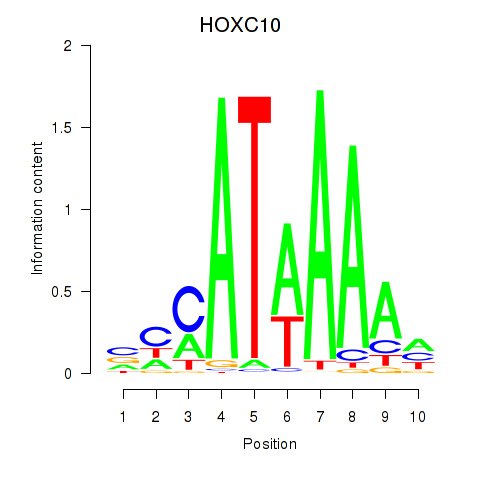
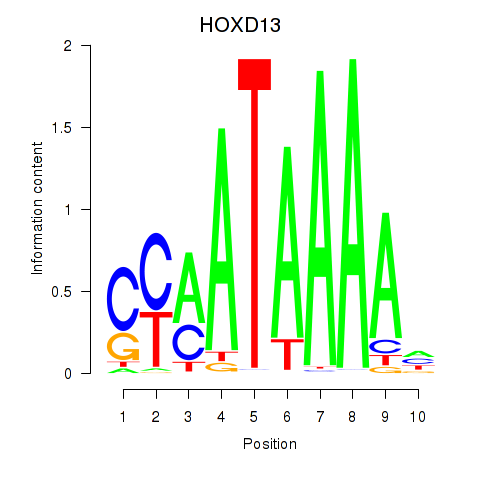
Results for HOXC10_HOXD13
Z-value: 0.85
Transcription factors associated with HOXC10_HOXD13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC10
|
ENSG00000180818.4 | homeobox C10 |
|
HOXD13
|
ENSG00000128714.5 | homeobox D13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD13 | hg19_v2_chr2_+_176957619_176957619 | 0.86 | 1.4e-01 | Click! |
| HOXC10 | hg19_v2_chr12_+_54378849_54378884 | -0.85 | 1.5e-01 | Click! |
Activity profile of HOXC10_HOXD13 motif
Sorted Z-values of HOXC10_HOXD13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_46690839 | 1.17 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr8_+_52730143 | 0.60 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr4_+_124571409 | 0.58 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr19_+_35773242 | 0.53 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chr2_+_200472779 | 0.52 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chr17_+_22022437 | 0.43 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr1_+_116654376 | 0.41 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr2_+_109204909 | 0.40 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr4_-_39033963 | 0.37 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr10_+_35456444 | 0.36 |
ENST00000361599.4
|
CREM
|
cAMP responsive element modulator |
| chr15_+_63414760 | 0.36 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr18_+_3252265 | 0.35 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr12_+_54384370 | 0.33 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr9_+_82188077 | 0.32 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr5_+_136070614 | 0.32 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr18_+_68002675 | 0.31 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr7_-_75401513 | 0.31 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr8_+_26150628 | 0.31 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr16_+_16429787 | 0.29 |
ENST00000331436.4
ENST00000541593.1 |
AC138969.4
|
Protein PKD1P1 |
| chr8_+_76452097 | 0.29 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr16_+_16472912 | 0.27 |
ENST00000530217.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr20_+_16729003 | 0.26 |
ENST00000246081.2
|
OTOR
|
otoraplin |
| chr14_-_51027838 | 0.25 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr7_+_108210012 | 0.24 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr4_+_39046615 | 0.24 |
ENST00000261425.3
ENST00000508137.2 |
KLHL5
|
kelch-like family member 5 |
| chr8_-_73793975 | 0.24 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr14_-_69444957 | 0.23 |
ENST00000556571.1
ENST00000553659.1 ENST00000555616.1 |
ACTN1
|
actinin, alpha 1 |
| chr13_-_44735393 | 0.22 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr15_-_71184724 | 0.21 |
ENST00000560604.1
|
THAP10
|
THAP domain containing 10 |
| chr5_+_95997918 | 0.21 |
ENST00000395812.2
ENST00000395813.1 ENST00000359176.4 ENST00000325674.7 |
CAST
|
calpastatin |
| chr15_-_80263506 | 0.21 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr14_-_57960456 | 0.21 |
ENST00000534126.1
ENST00000422976.2 |
C14orf105
|
chromosome 14 open reading frame 105 |
| chr12_-_112443830 | 0.20 |
ENST00000550037.1
ENST00000549425.1 |
TMEM116
|
transmembrane protein 116 |
| chr14_+_104710541 | 0.20 |
ENST00000419115.1
|
C14orf144
|
chromosome 14 open reading frame 144 |
| chr17_-_48943706 | 0.20 |
ENST00000499247.2
|
TOB1
|
transducer of ERBB2, 1 |
| chr7_-_27205136 | 0.20 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chr2_+_183580954 | 0.20 |
ENST00000264065.7
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr3_+_159557637 | 0.19 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr12_+_80838126 | 0.18 |
ENST00000266688.5
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q |
| chr14_+_39944025 | 0.18 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr7_-_108209897 | 0.18 |
ENST00000313516.5
|
THAP5
|
THAP domain containing 5 |
| chr13_+_108921977 | 0.18 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr18_-_74728998 | 0.18 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr2_+_109204743 | 0.18 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr4_-_48018680 | 0.18 |
ENST00000513178.1
|
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr13_-_31736132 | 0.18 |
ENST00000429785.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr14_-_55369525 | 0.17 |
ENST00000543643.2
ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1
|
GTP cyclohydrolase 1 |
| chr18_-_12656715 | 0.17 |
ENST00000462226.1
ENST00000497844.2 ENST00000309836.5 ENST00000453447.2 |
SPIRE1
|
spire-type actin nucleation factor 1 |
| chr10_+_21823079 | 0.17 |
ENST00000377100.3
ENST00000377072.3 ENST00000446906.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr1_-_150669604 | 0.17 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr16_+_333152 | 0.17 |
ENST00000219406.6
ENST00000404312.1 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chr12_+_52695617 | 0.16 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr9_-_16705069 | 0.16 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr2_-_69180083 | 0.16 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr6_+_76599809 | 0.16 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr8_+_38831683 | 0.16 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chrX_-_19688475 | 0.16 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr12_-_100486668 | 0.16 |
ENST00000550544.1
ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L
|
UHRF1 binding protein 1-like |
| chr9_+_108463234 | 0.15 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr16_+_14805546 | 0.15 |
ENST00000552140.1
|
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr15_+_49715293 | 0.15 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr3_+_142342228 | 0.15 |
ENST00000337777.3
|
PLS1
|
plastin 1 |
| chr11_+_101785727 | 0.15 |
ENST00000263468.8
|
KIAA1377
|
KIAA1377 |
| chr7_+_134464414 | 0.15 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr18_-_74839891 | 0.15 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr1_+_84630645 | 0.15 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr5_-_79946820 | 0.15 |
ENST00000604882.1
|
MTRNR2L2
|
MT-RNR2-like 2 |
| chr17_-_62499334 | 0.15 |
ENST00000579996.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr13_-_31736478 | 0.14 |
ENST00000445273.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr12_-_95510743 | 0.14 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr15_+_71228826 | 0.14 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr12_+_110718428 | 0.14 |
ENST00000552636.1
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr7_+_77428149 | 0.14 |
ENST00000415251.2
ENST00000275575.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr1_-_86848760 | 0.14 |
ENST00000460698.2
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr7_-_69062391 | 0.14 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr4_-_103746683 | 0.14 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_-_27114577 | 0.14 |
ENST00000356950.1
ENST00000396891.4 |
HIST1H2BK
|
histone cluster 1, H2bk |
| chr16_-_18466642 | 0.14 |
ENST00000541810.1
ENST00000531453.2 |
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr3_-_25915189 | 0.13 |
ENST00000451284.1
|
LINC00692
|
long intergenic non-protein coding RNA 692 |
| chr12_+_20963647 | 0.13 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr19_+_36706024 | 0.13 |
ENST00000443387.2
|
ZNF146
|
zinc finger protein 146 |
| chrX_+_73164149 | 0.13 |
ENST00000602938.1
ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr5_+_95997769 | 0.13 |
ENST00000338252.3
ENST00000508830.1 |
CAST
|
calpastatin |
| chr1_+_62439037 | 0.13 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr13_+_78315528 | 0.13 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr1_+_229440129 | 0.13 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr2_-_74555350 | 0.13 |
ENST00000444570.1
|
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chrX_+_154444643 | 0.13 |
ENST00000286428.5
|
VBP1
|
von Hippel-Lindau binding protein 1 |
| chr8_+_92114060 | 0.13 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr16_-_24942411 | 0.13 |
ENST00000571843.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr19_-_39303576 | 0.13 |
ENST00000594209.1
|
LGALS4
|
lectin, galactoside-binding, soluble, 4 |
| chr9_-_37384431 | 0.13 |
ENST00000452923.1
|
RP11-397D12.4
|
RP11-397D12.4 |
| chr2_+_67624430 | 0.12 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr4_-_103746924 | 0.12 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr5_-_36152031 | 0.12 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr15_+_55700741 | 0.12 |
ENST00000569691.1
|
C15orf65
|
chromosome 15 open reading frame 65 |
| chr18_+_21032781 | 0.12 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chrX_-_139866723 | 0.12 |
ENST00000370532.2
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa |
| chr1_-_154178803 | 0.12 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr2_+_64073187 | 0.12 |
ENST00000491621.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr12_-_76461249 | 0.12 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr9_+_130478345 | 0.12 |
ENST00000373289.3
ENST00000393748.4 |
TTC16
|
tetratricopeptide repeat domain 16 |
| chr3_+_142342240 | 0.12 |
ENST00000497199.1
|
PLS1
|
plastin 1 |
| chr4_-_103747011 | 0.12 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr14_+_93357749 | 0.12 |
ENST00000557689.1
|
RP11-862G15.1
|
RP11-862G15.1 |
| chr6_-_30080863 | 0.12 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chr12_-_76477707 | 0.12 |
ENST00000551992.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr1_+_151735431 | 0.12 |
ENST00000321531.5
ENST00000315067.8 |
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr1_-_9129631 | 0.12 |
ENST00000377414.3
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr13_-_31736027 | 0.12 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr5_+_43121698 | 0.12 |
ENST00000505606.2
ENST00000509634.1 ENST00000509341.1 |
ZNF131
|
zinc finger protein 131 |
| chr3_+_15045419 | 0.11 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr12_-_18243075 | 0.11 |
ENST00000536890.1
|
RERGL
|
RERG/RAS-like |
| chr12_+_6309517 | 0.11 |
ENST00000382519.4
ENST00000009180.4 |
CD9
|
CD9 molecule |
| chr1_+_28527070 | 0.11 |
ENST00000596102.1
|
AL353354.2
|
AL353354.2 |
| chr2_+_207024306 | 0.11 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr17_-_39526052 | 0.11 |
ENST00000251646.3
|
KRT33B
|
keratin 33B |
| chr11_+_102217936 | 0.11 |
ENST00000532832.1
ENST00000530675.1 ENST00000533742.1 ENST00000227758.2 ENST00000532672.1 ENST00000531259.1 ENST00000527465.1 |
BIRC2
|
baculoviral IAP repeat containing 2 |
| chr19_+_30433372 | 0.11 |
ENST00000312051.6
|
URI1
|
URI1, prefoldin-like chaperone |
| chr5_-_137475071 | 0.11 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr20_+_37554955 | 0.11 |
ENST00000217429.4
|
FAM83D
|
family with sequence similarity 83, member D |
| chr8_+_86089619 | 0.11 |
ENST00000256117.5
ENST00000416274.2 |
E2F5
|
E2F transcription factor 5, p130-binding |
| chr21_+_25801041 | 0.11 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr4_-_52883786 | 0.11 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr2_-_207023918 | 0.11 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr18_+_20494078 | 0.11 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr4_+_86525299 | 0.11 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr2_-_136288113 | 0.11 |
ENST00000401392.1
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr2_-_153032484 | 0.11 |
ENST00000263904.4
|
STAM2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
| chr18_-_33709268 | 0.11 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr10_+_85933494 | 0.11 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr7_-_50860565 | 0.11 |
ENST00000403097.1
|
GRB10
|
growth factor receptor-bound protein 10 |
| chr22_-_44708731 | 0.10 |
ENST00000381176.4
|
KIAA1644
|
KIAA1644 |
| chr9_+_130911770 | 0.10 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr5_-_145562147 | 0.10 |
ENST00000545646.1
ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS
|
leucyl-tRNA synthetase |
| chr4_-_170897045 | 0.10 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr18_-_37380230 | 0.10 |
ENST00000591629.1
|
LINC00669
|
long intergenic non-protein coding RNA 669 |
| chr7_+_77428066 | 0.10 |
ENST00000422959.2
ENST00000307305.8 ENST00000424760.1 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr1_+_209859510 | 0.10 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr3_+_57875738 | 0.10 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chrM_+_12331 | 0.10 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr11_-_27723158 | 0.10 |
ENST00000395980.2
|
BDNF
|
brain-derived neurotrophic factor |
| chr10_-_14050522 | 0.10 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr15_+_41549105 | 0.10 |
ENST00000560965.1
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr11_-_76155618 | 0.10 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr12_+_45686457 | 0.10 |
ENST00000441606.2
|
ANO6
|
anoctamin 6 |
| chr9_-_21305312 | 0.10 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr5_+_167913450 | 0.10 |
ENST00000231572.3
ENST00000538719.1 |
RARS
|
arginyl-tRNA synthetase |
| chr17_+_61473104 | 0.10 |
ENST00000583016.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr1_-_161207986 | 0.10 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr4_-_130692631 | 0.10 |
ENST00000500092.2
ENST00000509105.1 |
RP11-519M16.1
|
RP11-519M16.1 |
| chr14_-_50583271 | 0.10 |
ENST00000395860.2
ENST00000395859.2 |
VCPKMT
|
valosin containing protein lysine (K) methyltransferase |
| chr12_-_52800139 | 0.10 |
ENST00000257974.2
|
KRT82
|
keratin 82 |
| chr11_-_31531121 | 0.10 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr3_-_121379739 | 0.10 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr2_-_157198860 | 0.10 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr1_-_100643765 | 0.10 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr17_+_65713925 | 0.10 |
ENST00000253247.4
|
NOL11
|
nucleolar protein 11 |
| chr11_+_112047087 | 0.10 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr17_-_62340581 | 0.10 |
ENST00000258991.3
ENST00000583738.1 ENST00000584379.1 |
TEX2
|
testis expressed 2 |
| chr5_+_94890840 | 0.10 |
ENST00000504763.1
|
ARSK
|
arylsulfatase family, member K |
| chr10_+_65281123 | 0.09 |
ENST00000298249.4
ENST00000373758.4 |
REEP3
|
receptor accessory protein 3 |
| chr11_+_94706804 | 0.09 |
ENST00000335080.5
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr15_+_85523671 | 0.09 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chr4_+_48018781 | 0.09 |
ENST00000295461.5
|
NIPAL1
|
NIPA-like domain containing 1 |
| chr10_-_14596140 | 0.09 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr7_-_92747269 | 0.09 |
ENST00000446617.1
ENST00000379958.2 |
SAMD9
|
sterile alpha motif domain containing 9 |
| chr14_+_62164340 | 0.09 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr5_+_32585605 | 0.09 |
ENST00000265073.4
ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr4_+_169013666 | 0.09 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr3_+_122296465 | 0.09 |
ENST00000483793.1
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15 |
| chr2_-_177684007 | 0.09 |
ENST00000451851.1
|
AC092162.1
|
AC092162.1 |
| chr16_-_15472151 | 0.09 |
ENST00000360151.4
ENST00000543801.1 |
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr17_-_47786375 | 0.09 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr7_+_35756092 | 0.09 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chr2_-_37374876 | 0.09 |
ENST00000405334.1
|
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chrX_+_73164167 | 0.09 |
ENST00000414209.1
ENST00000602895.1 ENST00000453317.1 ENST00000602546.1 ENST00000602985.1 ENST00000415215.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr6_-_75828774 | 0.09 |
ENST00000493109.2
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr1_-_150669500 | 0.09 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr5_+_39105358 | 0.09 |
ENST00000593965.1
|
AC008964.1
|
AC008964.1 |
| chr6_-_10115007 | 0.09 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr1_-_151148492 | 0.09 |
ENST00000295314.4
|
TMOD4
|
tropomodulin 4 (muscle) |
| chr20_+_19870167 | 0.09 |
ENST00000440354.2
|
RIN2
|
Ras and Rab interactor 2 |
| chr21_-_35014027 | 0.09 |
ENST00000399442.1
ENST00000413017.2 ENST00000445393.1 ENST00000417979.1 ENST00000426935.1 ENST00000381540.3 ENST00000361534.2 ENST00000381554.3 |
CRYZL1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr17_+_41150479 | 0.09 |
ENST00000589913.1
|
RPL27
|
ribosomal protein L27 |
| chr14_+_73563735 | 0.09 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr16_+_25123148 | 0.09 |
ENST00000570981.1
|
LCMT1
|
leucine carboxyl methyltransferase 1 |
| chr2_-_136678123 | 0.09 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr16_-_67978016 | 0.09 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr11_+_113185778 | 0.09 |
ENST00000524580.2
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr1_+_52082751 | 0.09 |
ENST00000447887.1
ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr4_+_42399856 | 0.09 |
ENST00000319234.4
|
SHISA3
|
shisa family member 3 |
| chr3_-_195310802 | 0.09 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr11_-_61196858 | 0.09 |
ENST00000413184.2
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr2_-_119605253 | 0.09 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr8_+_125486939 | 0.09 |
ENST00000303545.3
|
RNF139
|
ring finger protein 139 |
| chr7_+_22766766 | 0.09 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr2_-_151344172 | 0.09 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr11_+_69924639 | 0.08 |
ENST00000538023.1
ENST00000398543.2 |
ANO1
|
anoctamin 1, calcium activated chloride channel |
| chr17_-_41132088 | 0.08 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC10_HOXD13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.1 | 0.3 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.2 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.4 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 0.2 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.0 | 1.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.1 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.4 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.2 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.1 | GO:1902683 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.2 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0072615 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.4 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.3 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.0 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:1902033 | regulation of hematopoietic stem cell proliferation(GO:1902033) |
| 0.0 | 0.2 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.0 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 0.1 | GO:0021557 | optic cup structural organization(GO:0003409) oculomotor nerve development(GO:0021557) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.0 | 0.1 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:0070885 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.0 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.1 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.0 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.2 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.2 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.1 | GO:0086039 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.1 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.0 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.0 | GO:0071209 | U7 snRNA binding(GO:0071209) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |