Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
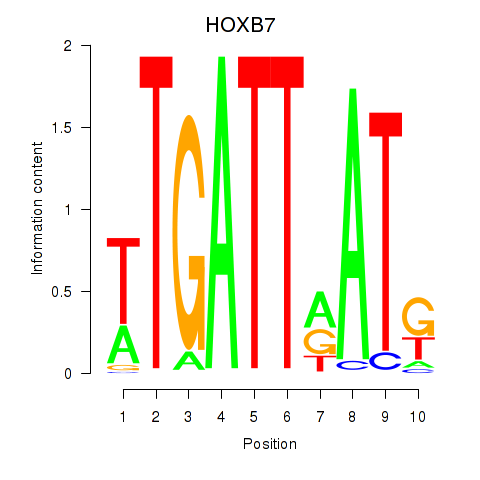
Results for HOXB7
Z-value: 0.48
Transcription factors associated with HOXB7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB7
|
ENSG00000260027.3 | homeobox B7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB7 | hg19_v2_chr17_-_46688334_46688385 | 0.39 | 6.1e-01 | Click! |
Activity profile of HOXB7 motif
Sorted Z-values of HOXB7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_20187174 | 0.23 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr18_+_44526786 | 0.19 |
ENST00000245121.5
ENST00000356157.7 |
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr10_-_18948208 | 0.19 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr1_+_112016414 | 0.19 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr11_+_28724129 | 0.19 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr17_+_70036164 | 0.17 |
ENST00000602013.1
|
AC007461.1
|
Uncharacterized protein |
| chr9_-_75488984 | 0.17 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr4_+_26323764 | 0.15 |
ENST00000514730.1
ENST00000507574.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr9_-_15472730 | 0.15 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr12_-_91546926 | 0.15 |
ENST00000550758.1
|
DCN
|
decorin |
| chr4_+_26324474 | 0.15 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr8_-_91618285 | 0.15 |
ENST00000517505.1
|
LINC01030
|
long intergenic non-protein coding RNA 1030 |
| chr17_+_7758374 | 0.14 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr9_-_85882145 | 0.14 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr12_-_95510743 | 0.14 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr14_+_45464658 | 0.14 |
ENST00000555874.1
|
FAM179B
|
family with sequence similarity 179, member B |
| chr3_+_159943362 | 0.13 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr11_-_61196858 | 0.13 |
ENST00000413184.2
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr3_+_52570610 | 0.13 |
ENST00000307106.3
ENST00000477703.1 ENST00000476842.1 |
SMIM4
|
small integral membrane protein 4 |
| chr13_-_48612067 | 0.12 |
ENST00000543413.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr12_-_54653313 | 0.12 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr19_+_20959098 | 0.11 |
ENST00000360204.5
ENST00000594534.1 |
ZNF66
|
zinc finger protein 66 |
| chr5_-_68339648 | 0.11 |
ENST00000479830.2
|
CTC-498J12.1
|
CTC-498J12.1 |
| chr3_+_157827841 | 0.11 |
ENST00000295930.3
ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr8_-_6115044 | 0.11 |
ENST00000519555.1
|
RP11-124B13.1
|
RP11-124B13.1 |
| chr18_+_44526744 | 0.11 |
ENST00000585469.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr2_+_44001172 | 0.10 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr6_-_138833630 | 0.10 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr17_+_58018269 | 0.10 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr11_-_8795787 | 0.10 |
ENST00000528196.1
ENST00000533681.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr5_-_148758839 | 0.10 |
ENST00000261796.3
|
IL17B
|
interleukin 17B |
| chr17_-_38821373 | 0.10 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr11_+_6947647 | 0.10 |
ENST00000278319.5
|
ZNF215
|
zinc finger protein 215 |
| chr3_-_71114066 | 0.10 |
ENST00000485326.2
|
FOXP1
|
forkhead box P1 |
| chr12_-_118628350 | 0.10 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr15_+_49715449 | 0.10 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr12_-_91573132 | 0.09 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr11_+_31833939 | 0.09 |
ENST00000530348.1
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr17_+_61151306 | 0.09 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr21_+_41029235 | 0.09 |
ENST00000380618.1
|
B3GALT5
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 |
| chr14_-_34931458 | 0.09 |
ENST00000298130.4
|
SPTSSA
|
serine palmitoyltransferase, small subunit A |
| chr16_+_66442411 | 0.09 |
ENST00000499966.1
|
LINC00920
|
long intergenic non-protein coding RNA 920 |
| chr20_+_10199566 | 0.09 |
ENST00000430336.1
|
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr19_+_44645700 | 0.09 |
ENST00000592437.1
|
ZNF234
|
zinc finger protein 234 |
| chr10_+_47894572 | 0.09 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr2_+_63277927 | 0.08 |
ENST00000282549.2
|
OTX1
|
orthodenticle homeobox 1 |
| chr9_+_104161123 | 0.08 |
ENST00000374861.3
ENST00000339664.2 ENST00000259395.4 |
ZNF189
|
zinc finger protein 189 |
| chr5_-_38557561 | 0.08 |
ENST00000511561.1
|
LIFR
|
leukemia inhibitory factor receptor alpha |
| chr12_-_15104040 | 0.08 |
ENST00000541644.1
ENST00000545895.1 |
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr17_-_49021974 | 0.08 |
ENST00000501718.2
|
RP11-700H6.1
|
RP11-700H6.1 |
| chr1_-_200589859 | 0.08 |
ENST00000367350.4
|
KIF14
|
kinesin family member 14 |
| chr11_-_96123022 | 0.08 |
ENST00000542662.1
|
CCDC82
|
coiled-coil domain containing 82 |
| chr2_-_230787879 | 0.08 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr3_+_141105705 | 0.08 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr15_+_82722225 | 0.08 |
ENST00000300515.8
|
GOLGA6L9
|
golgin A6 family-like 9 |
| chr8_+_56685701 | 0.08 |
ENST00000260129.5
|
TGS1
|
trimethylguanosine synthase 1 |
| chr1_+_168148273 | 0.07 |
ENST00000367830.3
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr15_-_30706365 | 0.07 |
ENST00000327271.10
ENST00000544495.1 |
GOLGA8R
|
golgin A8 family, member R |
| chr8_+_9953061 | 0.07 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr8_-_42360015 | 0.07 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr5_-_179047881 | 0.07 |
ENST00000521173.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr6_+_27925019 | 0.07 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr11_+_57365150 | 0.07 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr10_+_47894023 | 0.07 |
ENST00000358474.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr2_+_27886330 | 0.07 |
ENST00000326019.6
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr14_+_56078695 | 0.07 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr3_-_93692781 | 0.07 |
ENST00000394236.3
|
PROS1
|
protein S (alpha) |
| chr5_-_132073111 | 0.07 |
ENST00000403231.1
|
KIF3A
|
kinesin family member 3A |
| chr10_+_50887683 | 0.07 |
ENST00000374113.3
ENST00000374111.3 ENST00000374112.3 ENST00000535836.1 |
C10orf53
|
chromosome 10 open reading frame 53 |
| chr8_-_99955042 | 0.07 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr11_-_57194817 | 0.07 |
ENST00000529748.1
ENST00000525474.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chr5_-_179045199 | 0.07 |
ENST00000523921.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr15_+_58724184 | 0.07 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr10_+_13142225 | 0.07 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr19_+_56270380 | 0.06 |
ENST00000434937.2
|
RFPL4A
|
ret finger protein-like 4A |
| chr2_+_234104079 | 0.06 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr14_-_101295407 | 0.06 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr20_+_5931497 | 0.06 |
ENST00000378886.2
ENST00000265187.4 |
MCM8
|
minichromosome maintenance complex component 8 |
| chr15_+_86098670 | 0.06 |
ENST00000558811.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr8_-_109799793 | 0.06 |
ENST00000297459.3
|
TMEM74
|
transmembrane protein 74 |
| chr10_+_124320156 | 0.06 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr17_+_67498538 | 0.06 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr15_+_23255242 | 0.06 |
ENST00000450802.3
|
GOLGA8I
|
golgin A8 family, member I |
| chr22_+_46449674 | 0.06 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr7_-_107883678 | 0.06 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr4_-_177116772 | 0.06 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr3_-_141747950 | 0.06 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr12_+_21679220 | 0.06 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr15_+_30375158 | 0.06 |
ENST00000341650.6
ENST00000567927.1 |
GOLGA8J
|
golgin A8 family, member J |
| chr4_+_148653206 | 0.06 |
ENST00000336498.3
|
ARHGAP10
|
Rho GTPase activating protein 10 |
| chr8_-_93978216 | 0.06 |
ENST00000517751.1
ENST00000524107.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr8_+_91233711 | 0.06 |
ENST00000523283.1
ENST00000517400.1 |
LINC00534
|
long intergenic non-protein coding RNA 534 |
| chr12_+_100897130 | 0.06 |
ENST00000551379.1
ENST00000188403.7 ENST00000551184.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr1_-_91487770 | 0.06 |
ENST00000337393.5
|
ZNF644
|
zinc finger protein 644 |
| chr12_-_102455846 | 0.05 |
ENST00000545679.1
|
CCDC53
|
coiled-coil domain containing 53 |
| chr4_-_140005443 | 0.05 |
ENST00000510408.1
ENST00000420916.2 ENST00000358635.3 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr4_-_123542224 | 0.05 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr11_-_67981046 | 0.05 |
ENST00000402789.1
ENST00000402185.2 ENST00000458496.1 |
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr2_+_201242715 | 0.05 |
ENST00000421573.1
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr1_+_244227632 | 0.05 |
ENST00000598000.1
|
AL590483.1
|
Uncharacterized protein; cDNA FLJ42623 fis, clone BRACE3015894 |
| chr19_+_32836499 | 0.05 |
ENST00000311921.4
ENST00000544431.1 ENST00000355898.5 |
ZNF507
|
zinc finger protein 507 |
| chr12_-_102455902 | 0.05 |
ENST00000240079.6
|
CCDC53
|
coiled-coil domain containing 53 |
| chrX_+_48687283 | 0.05 |
ENST00000338270.1
|
ERAS
|
ES cell expressed Ras |
| chr1_-_91487806 | 0.05 |
ENST00000361321.5
|
ZNF644
|
zinc finger protein 644 |
| chr8_+_107593198 | 0.05 |
ENST00000517686.1
|
OXR1
|
oxidation resistance 1 |
| chr12_-_28125638 | 0.05 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chr14_+_62229075 | 0.05 |
ENST00000216294.4
|
SNAPC1
|
small nuclear RNA activating complex, polypeptide 1, 43kDa |
| chr11_+_107650219 | 0.05 |
ENST00000398067.1
|
AP001024.1
|
Uncharacterized protein |
| chr3_+_141105235 | 0.05 |
ENST00000503809.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr16_+_82660635 | 0.05 |
ENST00000567445.1
ENST00000446376.2 |
CDH13
|
cadherin 13 |
| chr15_-_42343388 | 0.05 |
ENST00000399518.3
|
PLA2G4E
|
phospholipase A2, group IVE |
| chr2_+_169658928 | 0.05 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr13_-_41240717 | 0.05 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr8_-_110986918 | 0.05 |
ENST00000297404.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr18_+_21464737 | 0.05 |
ENST00000586751.1
|
LAMA3
|
laminin, alpha 3 |
| chr17_-_45266542 | 0.05 |
ENST00000531206.1
ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27
|
cell division cycle 27 |
| chrX_+_95939638 | 0.05 |
ENST00000373061.3
ENST00000373054.4 ENST00000355827.4 |
DIAPH2
|
diaphanous-related formin 2 |
| chr12_+_21168630 | 0.05 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr11_+_71927807 | 0.05 |
ENST00000298223.6
ENST00000454954.2 ENST00000541003.1 ENST00000539412.1 ENST00000536778.1 ENST00000535625.1 ENST00000321324.7 |
FOLR2
|
folate receptor 2 (fetal) |
| chr19_-_57656570 | 0.05 |
ENST00000269834.1
|
ZIM3
|
zinc finger, imprinted 3 |
| chr10_+_24755416 | 0.05 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr1_-_242612726 | 0.05 |
ENST00000459864.1
|
PLD5
|
phospholipase D family, member 5 |
| chr6_-_10115007 | 0.05 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr16_-_18462221 | 0.05 |
ENST00000528301.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr17_-_38956205 | 0.05 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr5_+_162887556 | 0.05 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr3_+_139063372 | 0.05 |
ENST00000478464.1
|
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr3_+_136581042 | 0.05 |
ENST00000288986.2
ENST00000481752.1 ENST00000491539.1 ENST00000485096.1 |
NCK1
|
NCK adaptor protein 1 |
| chr11_-_57194948 | 0.05 |
ENST00000533235.1
ENST00000526621.1 ENST00000352187.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chr2_-_203735484 | 0.05 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr2_-_56150910 | 0.04 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr1_+_75600567 | 0.04 |
ENST00000356261.3
|
LHX8
|
LIM homeobox 8 |
| chrM_+_9207 | 0.04 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chrX_+_95939711 | 0.04 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr2_-_159237472 | 0.04 |
ENST00000409187.1
|
CCDC148
|
coiled-coil domain containing 148 |
| chr10_-_17243579 | 0.04 |
ENST00000525762.1
ENST00000412821.3 ENST00000351358.4 ENST00000377766.5 ENST00000358282.7 ENST00000488990.1 ENST00000377799.3 |
TRDMT1
|
tRNA aspartic acid methyltransferase 1 |
| chr7_-_120498357 | 0.04 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr10_+_13141585 | 0.04 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr8_-_124749609 | 0.04 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr2_+_190541153 | 0.04 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr2_+_161993412 | 0.04 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr8_+_9953214 | 0.04 |
ENST00000382490.5
|
MSRA
|
methionine sulfoxide reductase A |
| chr12_+_96252706 | 0.04 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr14_+_97263641 | 0.04 |
ENST00000216639.3
|
VRK1
|
vaccinia related kinase 1 |
| chr11_+_96123158 | 0.04 |
ENST00000332349.4
ENST00000458427.1 |
JRKL
|
jerky homolog-like (mouse) |
| chr7_+_141695671 | 0.04 |
ENST00000497673.1
ENST00000475668.2 |
MGAM
|
maltase-glucoamylase (alpha-glucosidase) |
| chrX_+_92929192 | 0.04 |
ENST00000332647.4
|
FAM133A
|
family with sequence similarity 133, member A |
| chr10_-_65028938 | 0.04 |
ENST00000402544.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr5_+_138209688 | 0.04 |
ENST00000518381.1
|
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr4_-_70080449 | 0.04 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr4_+_113568207 | 0.04 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr1_-_53018654 | 0.04 |
ENST00000257177.4
ENST00000355809.4 ENST00000528642.1 ENST00000470626.1 ENST00000371544.3 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr15_-_38519066 | 0.04 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chr2_+_201242941 | 0.04 |
ENST00000449647.1
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr12_+_20968608 | 0.04 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr15_-_74374891 | 0.04 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family, member A |
| chr16_+_16434185 | 0.04 |
ENST00000524823.2
|
AC138969.4
|
Protein PKD1P1 |
| chrX_-_102757802 | 0.04 |
ENST00000372633.1
|
RAB40A
|
RAB40A, member RAS oncogene family |
| chrX_-_49121165 | 0.04 |
ENST00000376207.4
ENST00000376199.2 |
FOXP3
|
forkhead box P3 |
| chr9_+_40028620 | 0.04 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr12_+_22852791 | 0.04 |
ENST00000413794.2
|
RP11-114G22.1
|
RP11-114G22.1 |
| chr3_-_145878954 | 0.04 |
ENST00000282903.5
ENST00000360060.3 |
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr3_-_183145873 | 0.04 |
ENST00000447025.2
ENST00000414362.2 ENST00000328913.3 |
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr3_+_138340067 | 0.04 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr15_+_76352178 | 0.04 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chrX_+_55744228 | 0.04 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr12_+_113229737 | 0.04 |
ENST00000551052.1
ENST00000415485.3 |
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr16_-_51185172 | 0.04 |
ENST00000251020.4
|
SALL1
|
spalt-like transcription factor 1 |
| chr2_+_120436760 | 0.04 |
ENST00000445518.1
ENST00000409951.1 |
TMEM177
|
transmembrane protein 177 |
| chr13_+_21714913 | 0.04 |
ENST00000450573.1
ENST00000467636.1 |
SAP18
|
Sin3A-associated protein, 18kDa |
| chr11_-_57194919 | 0.04 |
ENST00000532795.1
|
SLC43A3
|
solute carrier family 43, member 3 |
| chr3_-_33686925 | 0.04 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr2_+_234580499 | 0.04 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr11_-_104769141 | 0.04 |
ENST00000508062.1
ENST00000422698.2 |
CASP12
|
caspase 12 (gene/pseudogene) |
| chr3_+_138340049 | 0.04 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr2_+_169659121 | 0.04 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr14_-_74417096 | 0.04 |
ENST00000286544.3
|
FAM161B
|
family with sequence similarity 161, member B |
| chr1_+_66820058 | 0.04 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr9_-_129885010 | 0.04 |
ENST00000373425.3
|
ANGPTL2
|
angiopoietin-like 2 |
| chr5_-_58295712 | 0.03 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr6_-_90024967 | 0.03 |
ENST00000602399.1
|
GABRR2
|
gamma-aminobutyric acid (GABA) A receptor, rho 2 |
| chr18_+_29171689 | 0.03 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr2_+_234600253 | 0.03 |
ENST00000373424.1
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr10_+_13142075 | 0.03 |
ENST00000378757.2
ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN
|
optineurin |
| chr15_+_83098710 | 0.03 |
ENST00000561062.1
ENST00000358583.3 |
GOLGA6L9
|
golgin A6 family-like 20 |
| chr19_+_49199209 | 0.03 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr15_-_23692381 | 0.03 |
ENST00000567107.1
ENST00000345070.5 ENST00000312015.5 |
GOLGA6L2
|
golgin A6 family-like 2 |
| chr12_+_81471816 | 0.03 |
ENST00000261206.3
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3 |
| chr14_+_51955831 | 0.03 |
ENST00000356218.4
|
FRMD6
|
FERM domain containing 6 |
| chr21_-_33984456 | 0.03 |
ENST00000431216.1
ENST00000553001.1 ENST00000440966.1 |
AP000275.65
C21orf59
|
Uncharacterized protein chromosome 21 open reading frame 59 |
| chr17_+_10600894 | 0.03 |
ENST00000379774.4
|
ADPRM
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr10_+_124320195 | 0.03 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr12_+_22852866 | 0.03 |
ENST00000536744.1
|
RP11-114G22.1
|
RP11-114G22.1 |
| chr6_+_90272488 | 0.03 |
ENST00000485637.1
ENST00000522705.1 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr2_+_102413726 | 0.03 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr11_-_32816156 | 0.03 |
ENST00000531481.1
ENST00000335185.5 |
CCDC73
|
coiled-coil domain containing 73 |
| chr10_-_106240032 | 0.03 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr2_+_177025619 | 0.03 |
ENST00000410016.1
|
HOXD3
|
homeobox D3 |
| chr1_-_40562908 | 0.03 |
ENST00000527311.2
ENST00000449045.2 ENST00000372779.4 |
PPT1
|
palmitoyl-protein thioesterase 1 |
| chr3_+_87276407 | 0.03 |
ENST00000471660.1
ENST00000263780.4 ENST00000494980.1 |
CHMP2B
|
charged multivesicular body protein 2B |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.3 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.1 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0038185 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.0 | GO:0032714 | regulation of tolerance induction dependent upon immune response(GO:0002652) positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.0 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.0 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.0 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |