Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
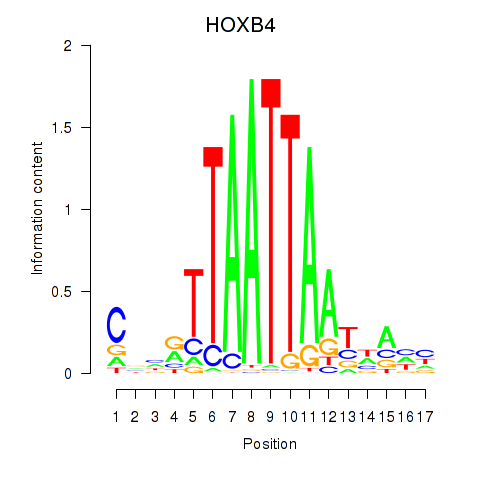
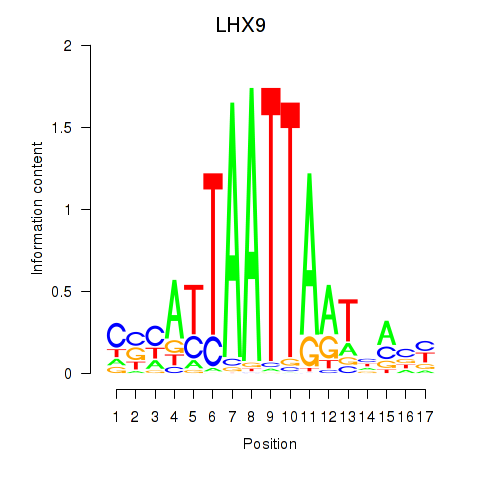
Results for HOXB4_LHX9
Z-value: 0.49
Transcription factors associated with HOXB4_LHX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB4
|
ENSG00000182742.5 | homeobox B4 |
|
LHX9
|
ENSG00000143355.11 | LIM homeobox 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX9 | hg19_v2_chr1_+_197881592_197881635 | 0.95 | 4.6e-02 | Click! |
| HOXB4 | hg19_v2_chr17_-_46657473_46657473 | -0.91 | 9.4e-02 | Click! |
Activity profile of HOXB4_LHX9 motif
Sorted Z-values of HOXB4_LHX9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_20187174 | 0.40 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr4_-_39033963 | 0.34 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr5_+_66300464 | 0.34 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr17_-_45266542 | 0.30 |
ENST00000531206.1
ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27
|
cell division cycle 27 |
| chr5_+_89770664 | 0.26 |
ENST00000503973.1
ENST00000399107.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr12_+_147052 | 0.25 |
ENST00000594563.1
|
AC026369.1
|
Uncharacterized protein |
| chrX_+_43515467 | 0.24 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chrX_+_108779004 | 0.22 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr5_+_89770696 | 0.21 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr5_-_89770582 | 0.21 |
ENST00000316610.6
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr5_+_179135246 | 0.20 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr5_+_136070614 | 0.20 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr19_+_56270380 | 0.19 |
ENST00000434937.2
|
RFPL4A
|
ret finger protein-like 4A |
| chr2_+_182850743 | 0.18 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr5_+_81575281 | 0.17 |
ENST00000380167.4
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr6_-_38670897 | 0.17 |
ENST00000373365.4
|
GLO1
|
glyoxalase I |
| chr12_+_64798095 | 0.17 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr12_-_10022735 | 0.17 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr2_-_74618907 | 0.16 |
ENST00000421392.1
ENST00000437375.1 |
DCTN1
|
dynactin 1 |
| chr17_+_1674982 | 0.16 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr17_+_20978854 | 0.16 |
ENST00000456235.1
|
AC087393.1
|
AC087393.1 |
| chr6_-_82957433 | 0.15 |
ENST00000306270.7
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr12_-_8803128 | 0.15 |
ENST00000543467.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr14_+_62164340 | 0.15 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr18_-_51751132 | 0.15 |
ENST00000256429.3
|
MBD2
|
methyl-CpG binding domain protein 2 |
| chr5_-_159546396 | 0.15 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chrX_+_100663243 | 0.14 |
ENST00000316594.5
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr6_-_138866823 | 0.14 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr8_-_93978216 | 0.14 |
ENST00000517751.1
ENST00000524107.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr18_-_32870148 | 0.14 |
ENST00000589178.1
ENST00000333206.5 ENST00000592278.1 ENST00000592211.1 ENST00000420878.3 ENST00000383091.2 ENST00000586922.2 |
ZSCAN30
RP11-158H5.7
|
zinc finger and SCAN domain containing 30 RP11-158H5.7 |
| chr2_+_182850551 | 0.14 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr8_-_1922789 | 0.14 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr2_-_27886676 | 0.14 |
ENST00000337768.5
|
SUPT7L
|
suppressor of Ty 7 (S. cerevisiae)-like |
| chr8_-_93978357 | 0.14 |
ENST00000522925.1
ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr8_-_93978309 | 0.14 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr18_-_74839891 | 0.14 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr7_+_102553430 | 0.13 |
ENST00000339431.4
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr14_-_51027838 | 0.13 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr8_-_93978346 | 0.13 |
ENST00000523580.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr8_-_90996459 | 0.13 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chr3_-_191000172 | 0.13 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr4_+_86525299 | 0.13 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr8_-_30670384 | 0.13 |
ENST00000221138.4
ENST00000518243.1 |
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr4_-_170948361 | 0.13 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chrX_+_13671225 | 0.13 |
ENST00000545566.1
ENST00000544987.1 ENST00000314720.4 |
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr2_+_29001711 | 0.13 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr4_+_76481258 | 0.12 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr14_+_56584414 | 0.12 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr16_+_82068585 | 0.12 |
ENST00000563491.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr3_-_48659193 | 0.12 |
ENST00000330862.3
|
TMEM89
|
transmembrane protein 89 |
| chr7_-_112635675 | 0.12 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr3_+_28390637 | 0.11 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr14_+_103851712 | 0.11 |
ENST00000440884.3
ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3
|
MAP/microtubule affinity-regulating kinase 3 |
| chr4_-_111120334 | 0.11 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr10_+_90660832 | 0.11 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr4_+_95128748 | 0.11 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr7_+_138145076 | 0.10 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr8_-_93978333 | 0.10 |
ENST00000524037.1
ENST00000520430.1 ENST00000521617.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr14_+_55494323 | 0.10 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr1_-_212965104 | 0.10 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr17_+_57233087 | 0.10 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chrX_-_71458802 | 0.10 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr4_+_108745711 | 0.10 |
ENST00000394684.4
|
SGMS2
|
sphingomyelin synthase 2 |
| chr14_-_39639523 | 0.10 |
ENST00000330149.5
ENST00000554018.1 ENST00000347691.5 |
TRAPPC6B
|
trafficking protein particle complex 6B |
| chr1_+_62439037 | 0.09 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr2_-_169887827 | 0.09 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr2_+_26624775 | 0.09 |
ENST00000288710.2
|
DRC1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr14_+_31046959 | 0.09 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr3_+_44840679 | 0.09 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr1_-_109584608 | 0.09 |
ENST00000400794.3
ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47
|
WD repeat domain 47 |
| chr14_+_55493920 | 0.09 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr3_+_139063372 | 0.09 |
ENST00000478464.1
|
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr14_+_45366518 | 0.09 |
ENST00000557112.1
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chrX_+_72783026 | 0.09 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr22_-_32767017 | 0.09 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr2_+_219125714 | 0.09 |
ENST00000522678.1
ENST00000519574.1 ENST00000521462.1 |
GPBAR1
|
G protein-coupled bile acid receptor 1 |
| chr1_+_212965170 | 0.08 |
ENST00000532324.1
ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3
|
TatD DNase domain containing 3 |
| chr18_+_47087390 | 0.08 |
ENST00000583083.1
|
LIPG
|
lipase, endothelial |
| chr4_-_103748880 | 0.08 |
ENST00000453744.2
ENST00000349311.8 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr14_+_24641820 | 0.08 |
ENST00000560501.1
|
REC8
|
REC8 meiotic recombination protein |
| chr18_+_20494078 | 0.08 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr15_-_72523924 | 0.08 |
ENST00000566809.1
ENST00000567087.1 ENST00000569050.1 ENST00000568883.1 |
PKM
|
pyruvate kinase, muscle |
| chr8_-_112248400 | 0.08 |
ENST00000519506.1
ENST00000522778.1 |
RP11-946L20.4
|
RP11-946L20.4 |
| chr17_+_66511540 | 0.08 |
ENST00000588188.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chrX_-_139047669 | 0.08 |
ENST00000370540.1
|
CXorf66
|
chromosome X open reading frame 66 |
| chr4_-_103997862 | 0.08 |
ENST00000394785.3
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr17_-_57229155 | 0.08 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr11_-_107729887 | 0.08 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr2_+_187454749 | 0.08 |
ENST00000261023.3
ENST00000374907.3 |
ITGAV
|
integrin, alpha V |
| chrX_+_84258832 | 0.07 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr22_-_32766972 | 0.07 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr6_+_28092338 | 0.07 |
ENST00000340487.4
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr7_+_116654935 | 0.07 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr14_-_45252031 | 0.07 |
ENST00000556405.1
|
RP11-398E10.1
|
RP11-398E10.1 |
| chr7_-_150020750 | 0.07 |
ENST00000539352.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr19_-_51920952 | 0.07 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr10_-_13043697 | 0.07 |
ENST00000378825.3
|
CCDC3
|
coiled-coil domain containing 3 |
| chr8_-_41166953 | 0.07 |
ENST00000220772.3
|
SFRP1
|
secreted frizzled-related protein 1 |
| chr1_-_77685084 | 0.07 |
ENST00000370812.3
ENST00000359130.1 ENST00000445065.1 ENST00000370813.5 |
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr12_-_74686314 | 0.07 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr4_-_140201333 | 0.07 |
ENST00000398955.1
|
MGARP
|
mitochondria-localized glutamic acid-rich protein |
| chr4_-_103998439 | 0.07 |
ENST00000503230.1
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr3_-_141747950 | 0.07 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chrX_+_107334895 | 0.07 |
ENST00000372232.3
ENST00000345734.3 ENST00000372254.3 |
ATG4A
|
autophagy related 4A, cysteine peptidase |
| chr1_+_155278625 | 0.07 |
ENST00000368356.4
ENST00000356657.6 |
FDPS
|
farnesyl diphosphate synthase |
| chr2_-_27886460 | 0.06 |
ENST00000404798.2
ENST00000405491.1 ENST00000464789.2 ENST00000406540.1 |
SUPT7L
|
suppressor of Ty 7 (S. cerevisiae)-like |
| chr15_+_94899183 | 0.06 |
ENST00000557742.1
|
MCTP2
|
multiple C2 domains, transmembrane 2 |
| chr7_-_150020814 | 0.06 |
ENST00000477871.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr2_+_103089756 | 0.06 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chrX_+_77166172 | 0.06 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr3_+_23851928 | 0.06 |
ENST00000467766.1
ENST00000424381.1 |
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr1_-_31666767 | 0.06 |
ENST00000530145.1
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr11_+_57480046 | 0.06 |
ENST00000378312.4
ENST00000278422.4 |
TMX2
|
thioredoxin-related transmembrane protein 2 |
| chr20_-_1447547 | 0.06 |
ENST00000476071.1
|
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr14_+_67831576 | 0.06 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr2_-_128284020 | 0.06 |
ENST00000295321.4
ENST00000455721.2 |
IWS1
|
IWS1 homolog (S. cerevisiae) |
| chr14_+_23938891 | 0.06 |
ENST00000408901.3
ENST00000397154.3 ENST00000555128.1 |
NGDN
|
neuroguidin, EIF4E binding protein |
| chr2_-_191115229 | 0.06 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr6_+_129204337 | 0.06 |
ENST00000421865.2
|
LAMA2
|
laminin, alpha 2 |
| chr4_+_113568207 | 0.06 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr15_+_80351910 | 0.06 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr11_-_46408107 | 0.06 |
ENST00000433765.2
|
CHRM4
|
cholinergic receptor, muscarinic 4 |
| chr15_+_64680003 | 0.06 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr17_+_33914424 | 0.06 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr4_+_56815102 | 0.06 |
ENST00000257287.4
|
CEP135
|
centrosomal protein 135kDa |
| chr4_+_157997273 | 0.06 |
ENST00000541722.1
ENST00000512619.1 |
GLRB
|
glycine receptor, beta |
| chr14_+_45366472 | 0.06 |
ENST00000325192.3
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr4_+_25162253 | 0.06 |
ENST00000512921.1
|
PI4K2B
|
phosphatidylinositol 4-kinase type 2 beta |
| chr11_+_65265141 | 0.06 |
ENST00000534336.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr5_-_36152031 | 0.06 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr4_-_103749105 | 0.05 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr17_-_38956205 | 0.05 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr1_+_155278539 | 0.05 |
ENST00000447866.1
|
FDPS
|
farnesyl diphosphate synthase |
| chr12_-_112123524 | 0.05 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr8_-_90996837 | 0.05 |
ENST00000519426.1
ENST00000265433.3 |
NBN
|
nibrin |
| chr3_-_145940126 | 0.05 |
ENST00000498625.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr4_-_185275104 | 0.05 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr4_+_11470867 | 0.05 |
ENST00000515343.1
|
RP11-281P23.1
|
RP11-281P23.1 |
| chr18_+_56806701 | 0.05 |
ENST00000587834.1
|
SEC11C
|
SEC11 homolog C (S. cerevisiae) |
| chr14_-_35591433 | 0.05 |
ENST00000261475.5
ENST00000555644.1 |
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr19_-_36304201 | 0.05 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr5_-_102898465 | 0.05 |
ENST00000507423.1
ENST00000230792.2 |
NUDT12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr11_-_13011081 | 0.05 |
ENST00000532541.1
ENST00000526388.1 ENST00000534477.1 ENST00000531402.1 ENST00000527945.1 ENST00000504230.2 |
LINC00958
|
long intergenic non-protein coding RNA 958 |
| chr7_+_23145884 | 0.05 |
ENST00000409689.1
ENST00000410047.1 |
KLHL7
|
kelch-like family member 7 |
| chr20_-_1447467 | 0.05 |
ENST00000353088.2
ENST00000350991.4 |
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr6_+_130339710 | 0.05 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr8_-_101571933 | 0.05 |
ENST00000520311.1
|
ANKRD46
|
ankyrin repeat domain 46 |
| chr9_-_26947220 | 0.05 |
ENST00000520884.1
|
PLAA
|
phospholipase A2-activating protein |
| chr7_-_44580861 | 0.05 |
ENST00000546276.1
ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1
|
NPC1-like 1 |
| chr4_-_103749313 | 0.05 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr20_+_18488137 | 0.05 |
ENST00000450074.1
ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr7_+_141438118 | 0.05 |
ENST00000265304.6
ENST00000498107.1 ENST00000467681.1 ENST00000465582.1 ENST00000463093.1 |
SSBP1
|
single-stranded DNA binding protein 1, mitochondrial |
| chr15_+_48483736 | 0.05 |
ENST00000417307.2
ENST00000559641.1 |
CTXN2
SLC12A1
|
cortexin 2 solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr16_+_15489629 | 0.05 |
ENST00000396385.3
|
MPV17L
|
MPV17 mitochondrial membrane protein-like |
| chr11_+_118398178 | 0.05 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chr7_+_115862858 | 0.05 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr11_+_59522837 | 0.05 |
ENST00000437946.2
|
STX3
|
syntaxin 3 |
| chr11_-_18548426 | 0.05 |
ENST00000357193.3
ENST00000536719.1 |
TSG101
|
tumor susceptibility 101 |
| chr9_-_99540328 | 0.05 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr14_-_35591156 | 0.05 |
ENST00000554361.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr3_+_111717511 | 0.05 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr11_+_35201826 | 0.05 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr10_+_695888 | 0.05 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr2_+_102615416 | 0.05 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr2_+_201242715 | 0.05 |
ENST00000421573.1
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr18_+_21033239 | 0.05 |
ENST00000581585.1
ENST00000577501.1 |
RIOK3
|
RIO kinase 3 |
| chr5_+_112849373 | 0.05 |
ENST00000161863.4
ENST00000515883.1 |
YTHDC2
|
YTH domain containing 2 |
| chrX_+_19362011 | 0.05 |
ENST00000379806.5
ENST00000545074.1 ENST00000540249.1 ENST00000423505.1 ENST00000417819.1 ENST00000422285.2 ENST00000355808.5 ENST00000379805.3 |
PDHA1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr4_+_80584903 | 0.05 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr3_+_138340067 | 0.05 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr1_+_146714291 | 0.05 |
ENST00000431239.1
ENST00000369259.3 ENST00000369258.4 ENST00000361293.5 |
CHD1L
|
chromodomain helicase DNA binding protein 1-like |
| chr19_+_54466179 | 0.05 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr10_+_6821545 | 0.05 |
ENST00000436383.1
|
LINC00707
|
long intergenic non-protein coding RNA 707 |
| chr17_-_38821373 | 0.05 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr5_-_125930929 | 0.05 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr2_+_234637754 | 0.04 |
ENST00000482026.1
ENST00000609767.1 |
UGT1A3
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chrX_-_102510045 | 0.04 |
ENST00000360000.4
|
TCEAL8
|
transcription elongation factor A (SII)-like 8 |
| chr14_+_55595762 | 0.04 |
ENST00000254301.9
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr16_+_24549014 | 0.04 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr12_+_32687221 | 0.04 |
ENST00000525053.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr19_-_14064114 | 0.04 |
ENST00000585607.1
ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1
|
podocan-like 1 |
| chr16_-_28608424 | 0.04 |
ENST00000335715.4
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr18_+_47087055 | 0.04 |
ENST00000577628.1
|
LIPG
|
lipase, endothelial |
| chr7_+_134528635 | 0.04 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr2_+_30670077 | 0.04 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr7_+_107224364 | 0.04 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr2_-_136678123 | 0.04 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr13_-_24007815 | 0.04 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr19_-_52307357 | 0.04 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr7_+_73868220 | 0.04 |
ENST00000455841.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr6_+_3259148 | 0.04 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr11_+_122753391 | 0.04 |
ENST00000307257.6
ENST00000227349.2 |
C11orf63
|
chromosome 11 open reading frame 63 |
| chr19_-_3772209 | 0.04 |
ENST00000555978.1
ENST00000555633.1 |
RAX2
|
retina and anterior neural fold homeobox 2 |
| chr3_-_151034734 | 0.04 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr2_+_27886330 | 0.04 |
ENST00000326019.6
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr10_-_27529486 | 0.04 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr3_+_157154578 | 0.04 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chrX_+_70503526 | 0.04 |
ENST00000413858.1
ENST00000450092.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr19_+_49199209 | 0.04 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chrM_+_9207 | 0.04 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB4_LHX9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.2 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:0045337 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.1 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.1 | GO:0090244 | negative regulation of B cell differentiation(GO:0045578) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.2 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.0 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.0 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.1 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.0 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.0 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.0 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |