Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for HOXB3
Z-value: 0.47
Transcription factors associated with HOXB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB3
|
ENSG00000120093.7 | homeobox B3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB3 | hg19_v2_chr17_-_46667594_46667619 | -0.43 | 5.7e-01 | Click! |
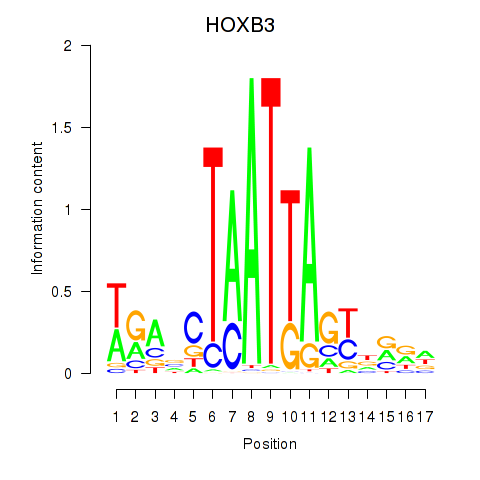
Activity profile of HOXB3 motif
Sorted Z-values of HOXB3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_26045603 | 0.49 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr17_+_62223320 | 0.41 |
ENST00000580828.1
ENST00000582965.1 |
SNORA76
|
small nucleolar RNA, H/ACA box 76 |
| chr7_-_99716914 | 0.39 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr21_-_27423339 | 0.25 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr7_+_100860949 | 0.20 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr9_+_131062367 | 0.20 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr17_+_74846230 | 0.19 |
ENST00000592919.1
|
LINC00868
|
long intergenic non-protein coding RNA 868 |
| chr8_+_39972170 | 0.17 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr7_-_73256838 | 0.17 |
ENST00000297873.4
|
WBSCR27
|
Williams Beuren syndrome chromosome region 27 |
| chr4_+_144354644 | 0.16 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr15_+_78830023 | 0.16 |
ENST00000599596.1
|
AC027228.1
|
HCG2005369; Uncharacterized protein |
| chr2_+_54350316 | 0.15 |
ENST00000606865.1
|
ACYP2
|
acylphosphatase 2, muscle type |
| chr2_-_236684438 | 0.15 |
ENST00000409661.1
|
AC064874.1
|
Uncharacterized protein |
| chr19_-_3557570 | 0.15 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr1_-_201140673 | 0.15 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr22_+_21996549 | 0.14 |
ENST00000248958.4
|
SDF2L1
|
stromal cell-derived factor 2-like 1 |
| chr13_-_30160925 | 0.14 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr20_+_43990576 | 0.14 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr6_+_26204825 | 0.13 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr2_+_191221240 | 0.13 |
ENST00000409027.1
ENST00000458193.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr12_+_78359999 | 0.13 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr17_-_73901494 | 0.12 |
ENST00000309352.3
|
MRPL38
|
mitochondrial ribosomal protein L38 |
| chr12_-_123201337 | 0.12 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr2_+_138722028 | 0.12 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chr14_+_74034310 | 0.12 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr2_+_200820494 | 0.12 |
ENST00000435773.2
|
C2orf47
|
chromosome 2 open reading frame 47 |
| chr14_+_32798547 | 0.12 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr5_+_140019004 | 0.11 |
ENST00000394671.3
ENST00000511410.1 ENST00000537378.1 |
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chr10_+_18549645 | 0.11 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr11_-_124981475 | 0.11 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr15_-_78526855 | 0.11 |
ENST00000541759.1
ENST00000558130.1 |
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr19_+_10362882 | 0.10 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr1_+_186265399 | 0.10 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr7_+_55980331 | 0.10 |
ENST00000429591.2
|
ZNF713
|
zinc finger protein 713 |
| chr7_-_100860851 | 0.10 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr16_+_28763108 | 0.10 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr1_+_16090914 | 0.10 |
ENST00000441801.2
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr14_+_101359265 | 0.10 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr4_+_40198527 | 0.10 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr2_+_196313239 | 0.10 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr19_+_48949030 | 0.09 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr14_+_56584414 | 0.09 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr9_-_35815013 | 0.09 |
ENST00000259667.5
|
HINT2
|
histidine triad nucleotide binding protein 2 |
| chr17_-_57229155 | 0.09 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr3_-_130745571 | 0.09 |
ENST00000514044.1
ENST00000264992.3 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr7_+_99202003 | 0.09 |
ENST00000609449.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr15_+_63050785 | 0.09 |
ENST00000472902.1
|
TLN2
|
talin 2 |
| chr5_+_140019079 | 0.09 |
ENST00000252100.6
|
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chr13_+_78315295 | 0.09 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr7_-_55620433 | 0.09 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr19_-_17366257 | 0.08 |
ENST00000594059.1
|
AC010646.3
|
Uncharacterized protein |
| chr15_+_92006567 | 0.08 |
ENST00000554333.1
|
RP11-661P17.1
|
RP11-661P17.1 |
| chr16_-_11036300 | 0.08 |
ENST00000331808.4
|
DEXI
|
Dexi homolog (mouse) |
| chr1_+_160370344 | 0.08 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr19_+_50270219 | 0.08 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr4_+_144312659 | 0.08 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr11_+_3819049 | 0.08 |
ENST00000396986.2
ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2
|
post-GPI attachment to proteins 2 |
| chr1_+_209602156 | 0.08 |
ENST00000429156.1
ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr20_-_44600810 | 0.08 |
ENST00000322927.2
ENST00000426788.1 |
ZNF335
|
zinc finger protein 335 |
| chr4_-_176733897 | 0.08 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr1_+_159750776 | 0.08 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr22_-_30960876 | 0.07 |
ENST00000401975.1
ENST00000428682.1 ENST00000423299.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr5_-_96478466 | 0.07 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr11_+_94706804 | 0.07 |
ENST00000335080.5
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr19_+_48949087 | 0.07 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr20_+_43991668 | 0.07 |
ENST00000243918.5
ENST00000426004.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr5_-_108063949 | 0.07 |
ENST00000606054.1
|
LINC01023
|
long intergenic non-protein coding RNA 1023 |
| chr7_+_74379083 | 0.07 |
ENST00000361825.7
|
GATSL1
|
GATS protein-like 1 |
| chr17_+_46970178 | 0.07 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr8_+_27169138 | 0.07 |
ENST00000522338.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr11_-_26593649 | 0.07 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr2_+_105050794 | 0.07 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr12_+_6419877 | 0.07 |
ENST00000536531.1
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr16_+_28648975 | 0.06 |
ENST00000529716.1
|
NPIPB8
|
nuclear pore complex interacting protein family, member B8 |
| chr6_+_150690028 | 0.06 |
ENST00000229447.5
ENST00000344419.3 |
IYD
|
iodotyrosine deiodinase |
| chr5_+_129083772 | 0.06 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr15_-_100273544 | 0.06 |
ENST00000409796.1
ENST00000545021.1 ENST00000344791.2 ENST00000332728.4 ENST00000450512.1 |
LYSMD4
|
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr1_-_51796226 | 0.06 |
ENST00000451380.1
ENST00000371747.3 ENST00000439482.1 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr3_-_121740969 | 0.06 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr4_-_40632881 | 0.06 |
ENST00000511598.1
|
RBM47
|
RNA binding motif protein 47 |
| chr12_+_56521840 | 0.06 |
ENST00000394048.5
|
ESYT1
|
extended synaptotagmin-like protein 1 |
| chrX_-_24690771 | 0.06 |
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr12_-_118796910 | 0.06 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr10_-_22292675 | 0.06 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr16_+_31724618 | 0.06 |
ENST00000530881.1
ENST00000529515.1 |
ZNF720
|
zinc finger protein 720 |
| chr7_+_90012986 | 0.06 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr16_-_31085514 | 0.06 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chr19_+_36632204 | 0.06 |
ENST00000592354.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr19_+_17638059 | 0.06 |
ENST00000599164.1
ENST00000449408.2 ENST00000600871.1 ENST00000599124.1 |
FAM129C
|
family with sequence similarity 129, member C |
| chr17_-_27418537 | 0.06 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr9_-_132383055 | 0.06 |
ENST00000372478.4
|
C9orf50
|
chromosome 9 open reading frame 50 |
| chr7_-_111032971 | 0.06 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr14_-_24711806 | 0.06 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr10_+_1205708 | 0.05 |
ENST00000425630.1
ENST00000583117.1 |
LINC00200
|
long intergenic non-protein coding RNA 200 |
| chr4_-_40632844 | 0.05 |
ENST00000505414.1
|
RBM47
|
RNA binding motif protein 47 |
| chr6_+_26501449 | 0.05 |
ENST00000244513.6
|
BTN1A1
|
butyrophilin, subfamily 1, member A1 |
| chr2_+_191208601 | 0.05 |
ENST00000413239.1
ENST00000431594.1 ENST00000444194.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr4_-_109541539 | 0.05 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr17_+_48624450 | 0.05 |
ENST00000006658.6
ENST00000356488.4 ENST00000393244.3 |
SPATA20
|
spermatogenesis associated 20 |
| chr2_-_220264703 | 0.05 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr4_-_100212132 | 0.05 |
ENST00000209668.2
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr9_+_5231413 | 0.05 |
ENST00000239316.4
|
INSL4
|
insulin-like 4 (placenta) |
| chr4_-_10686475 | 0.05 |
ENST00000226951.6
|
CLNK
|
cytokine-dependent hematopoietic cell linker |
| chr7_+_37723420 | 0.05 |
ENST00000476620.1
|
EPDR1
|
ependymin related 1 |
| chr2_+_234637754 | 0.05 |
ENST00000482026.1
ENST00000609767.1 |
UGT1A3
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr22_-_32766972 | 0.05 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr6_-_119256142 | 0.05 |
ENST00000425154.2
|
MCM9
|
minichromosome maintenance complex component 9 |
| chr2_+_166095898 | 0.05 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr1_-_68698222 | 0.05 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr1_-_1709845 | 0.05 |
ENST00000341426.5
ENST00000344463.4 |
NADK
|
NAD kinase |
| chr2_+_65215604 | 0.05 |
ENST00000531327.1
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr2_-_101925055 | 0.05 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr3_+_158519654 | 0.05 |
ENST00000415822.2
ENST00000392813.4 ENST00000264266.8 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr4_+_175204865 | 0.05 |
ENST00000505124.1
|
CEP44
|
centrosomal protein 44kDa |
| chr16_-_28374829 | 0.05 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chr1_+_144173162 | 0.05 |
ENST00000356801.6
|
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr15_-_55562582 | 0.05 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_+_128399965 | 0.05 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr19_-_3985455 | 0.05 |
ENST00000309311.6
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr2_-_112237835 | 0.05 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr10_+_5090940 | 0.04 |
ENST00000602997.1
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr12_+_94071341 | 0.04 |
ENST00000542893.2
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr12_+_128399917 | 0.04 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr12_-_8088773 | 0.04 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr12_-_23737534 | 0.04 |
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr11_+_33061543 | 0.04 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr10_-_135379132 | 0.04 |
ENST00000343131.5
|
SYCE1
|
synaptonemal complex central element protein 1 |
| chr9_+_130911723 | 0.04 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr1_-_54356404 | 0.04 |
ENST00000539954.1
|
YIPF1
|
Yip1 domain family, member 1 |
| chr19_-_55677999 | 0.04 |
ENST00000532817.1
ENST00000527223.2 ENST00000391720.4 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr12_+_56522001 | 0.04 |
ENST00000267113.4
ENST00000541590.1 |
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr19_-_55677920 | 0.04 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr17_-_73258425 | 0.04 |
ENST00000578348.1
ENST00000582486.1 ENST00000582717.1 |
GGA3
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr19_+_36632485 | 0.04 |
ENST00000586963.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr22_+_30792980 | 0.04 |
ENST00000403484.1
ENST00000405717.3 ENST00000402592.3 |
SEC14L2
|
SEC14-like 2 (S. cerevisiae) |
| chr20_-_5485166 | 0.04 |
ENST00000589201.1
ENST00000379053.4 |
LINC00654
|
long intergenic non-protein coding RNA 654 |
| chr1_-_21625486 | 0.04 |
ENST00000481130.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr2_+_234545092 | 0.04 |
ENST00000344644.5
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr16_-_28621312 | 0.04 |
ENST00000314752.7
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr3_+_40518599 | 0.04 |
ENST00000314686.5
ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619
|
zinc finger protein 619 |
| chr7_+_26331541 | 0.04 |
ENST00000416246.1
ENST00000338523.4 ENST00000412416.1 |
SNX10
|
sorting nexin 10 |
| chr14_-_23058063 | 0.04 |
ENST00000538631.1
ENST00000543337.1 ENST00000250498.4 |
DAD1
|
defender against cell death 1 |
| chr5_+_136070614 | 0.04 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr7_+_99647411 | 0.04 |
ENST00000438937.1
|
ZSCAN21
|
zinc finger and SCAN domain containing 21 |
| chr16_-_21868978 | 0.04 |
ENST00000357370.5
ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr14_-_24711865 | 0.04 |
ENST00000399423.4
ENST00000267415.7 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr19_-_46088068 | 0.04 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr7_-_99716952 | 0.04 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr19_+_10362577 | 0.03 |
ENST00000592514.1
ENST00000307422.5 ENST00000253099.6 ENST00000590150.1 ENST00000590669.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chrX_+_72783026 | 0.03 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr5_-_140027357 | 0.03 |
ENST00000252102.4
|
NDUFA2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| chrX_-_48824793 | 0.03 |
ENST00000376477.1
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr22_-_29107919 | 0.03 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr11_-_63439013 | 0.03 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chr6_+_34204642 | 0.03 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr14_+_57671888 | 0.03 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr19_+_17638041 | 0.03 |
ENST00000601861.1
|
FAM129C
|
family with sequence similarity 129, member C |
| chr19_-_6737576 | 0.03 |
ENST00000601716.1
ENST00000264080.7 |
GPR108
|
G protein-coupled receptor 108 |
| chr1_+_155006300 | 0.03 |
ENST00000295542.1
ENST00000392480.1 ENST00000423025.2 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr16_-_28621353 | 0.03 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr10_+_99627889 | 0.03 |
ENST00000596005.1
|
GOLGA7B
|
Golgin subfamily A member 7B; cDNA FLJ43465 fis, clone OCBBF2036476 |
| chr15_-_55562479 | 0.03 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_-_117175819 | 0.03 |
ENST00000261318.3
ENST00000536380.1 |
C12orf49
|
chromosome 12 open reading frame 49 |
| chr3_+_114012819 | 0.03 |
ENST00000383671.3
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr17_-_33446735 | 0.03 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr11_-_107729504 | 0.03 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr1_-_207226313 | 0.03 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr1_-_68698197 | 0.03 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr9_+_12775011 | 0.03 |
ENST00000319264.3
|
LURAP1L
|
leucine rich adaptor protein 1-like |
| chr1_+_158978768 | 0.03 |
ENST00000447473.2
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chrX_+_10031499 | 0.03 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr1_+_197237352 | 0.03 |
ENST00000538660.1
ENST00000367400.3 ENST00000367399.2 |
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr17_+_46970134 | 0.03 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr10_-_4285923 | 0.03 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr17_-_47786375 | 0.03 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr16_-_28937027 | 0.03 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr14_-_24711764 | 0.03 |
ENST00000557921.1
ENST00000558476.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr19_+_49990811 | 0.03 |
ENST00000391857.4
ENST00000467825.2 |
RPL13A
|
ribosomal protein L13a |
| chr16_+_31885079 | 0.03 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr19_+_9296279 | 0.03 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr19_+_11071546 | 0.03 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr11_+_94706973 | 0.03 |
ENST00000536741.1
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr16_-_30257122 | 0.03 |
ENST00000520915.1
|
RP11-347C12.1
|
Putative NPIP-like protein LOC613037 |
| chr8_-_41166953 | 0.02 |
ENST00000220772.3
|
SFRP1
|
secreted frizzled-related protein 1 |
| chr12_+_31812121 | 0.02 |
ENST00000395763.3
|
METTL20
|
methyltransferase like 20 |
| chr14_-_70883708 | 0.02 |
ENST00000256366.4
|
SYNJ2BP
|
synaptojanin 2 binding protein |
| chr1_-_1710229 | 0.02 |
ENST00000341991.3
|
NADK
|
NAD kinase |
| chr17_+_40610862 | 0.02 |
ENST00000393829.2
ENST00000546249.1 ENST00000537728.1 ENST00000264649.6 ENST00000585525.1 ENST00000343619.4 ENST00000544137.1 ENST00000589727.1 ENST00000587824.1 |
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1 |
| chr1_+_28261492 | 0.02 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr16_-_29415350 | 0.02 |
ENST00000524087.1
|
NPIPB11
|
nuclear pore complex interacting protein family, member B11 |
| chr11_-_60010556 | 0.02 |
ENST00000427611.2
|
MS4A4E
|
membrane-spanning 4-domains, subfamily A, member 4E |
| chr1_-_9953295 | 0.02 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr2_+_191208656 | 0.02 |
ENST00000458647.1
|
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr17_-_72772425 | 0.02 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr7_-_99717463 | 0.02 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr11_-_95522639 | 0.02 |
ENST00000536839.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr17_-_47785504 | 0.02 |
ENST00000514907.1
ENST00000503334.1 ENST00000508520.1 |
SLC35B1
|
solute carrier family 35, member B1 |
| chr9_-_3489406 | 0.02 |
ENST00000457373.1
|
RFX3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr15_+_64680003 | 0.02 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr17_+_46970127 | 0.02 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.0 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.0 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.0 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.0 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |