Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
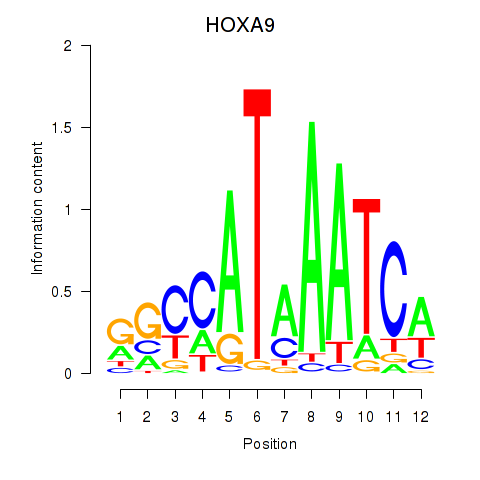
Results for HOXA9
Z-value: 0.52
Transcription factors associated with HOXA9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA9
|
ENSG00000078399.11 | homeobox A9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA9 | hg19_v2_chr7_-_27205136_27205164 | 0.71 | 2.9e-01 | Click! |
Activity profile of HOXA9 motif
Sorted Z-values of HOXA9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_15001430 | 0.43 |
ENST00000407572.1
|
MEIG1
|
meiosis/spermiogenesis associated 1 |
| chr18_+_1099004 | 0.35 |
ENST00000581556.1
|
RP11-78F17.1
|
RP11-78F17.1 |
| chr9_+_78505581 | 0.22 |
ENST00000376767.3
ENST00000376752.4 |
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr8_-_91618285 | 0.21 |
ENST00000517505.1
|
LINC01030
|
long intergenic non-protein coding RNA 1030 |
| chr15_+_63414760 | 0.20 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr12_-_121477039 | 0.20 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr12_-_13248598 | 0.20 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr4_+_76439095 | 0.19 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr5_+_158737824 | 0.18 |
ENST00000521472.1
|
AC008697.1
|
AC008697.1 |
| chr12_-_121476959 | 0.17 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr16_+_56691838 | 0.17 |
ENST00000394501.2
|
MT1F
|
metallothionein 1F |
| chr8_+_52730143 | 0.15 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr9_+_40028620 | 0.15 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr12_-_121476750 | 0.15 |
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr11_+_113185778 | 0.14 |
ENST00000524580.2
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr17_-_57229155 | 0.14 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr4_-_48018680 | 0.14 |
ENST00000513178.1
|
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr2_-_177684007 | 0.14 |
ENST00000451851.1
|
AC092162.1
|
AC092162.1 |
| chr1_-_43282945 | 0.14 |
ENST00000537227.1
|
CCDC23
|
coiled-coil domain containing 23 |
| chr5_-_168006324 | 0.14 |
ENST00000522176.1
|
PANK3
|
pantothenate kinase 3 |
| chr3_-_197686847 | 0.13 |
ENST00000265239.6
|
IQCG
|
IQ motif containing G |
| chr8_+_38261880 | 0.13 |
ENST00000527175.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr4_-_52883786 | 0.13 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr20_+_32250079 | 0.13 |
ENST00000375222.3
|
C20orf144
|
chromosome 20 open reading frame 144 |
| chr5_+_159436120 | 0.13 |
ENST00000522793.1
ENST00000231238.5 |
TTC1
|
tetratricopeptide repeat domain 1 |
| chr2_+_238600933 | 0.13 |
ENST00000420665.1
ENST00000392000.4 |
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1 |
| chr1_+_52082751 | 0.12 |
ENST00000447887.1
ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr4_+_39046615 | 0.12 |
ENST00000261425.3
ENST00000508137.2 |
KLHL5
|
kelch-like family member 5 |
| chr16_-_18462221 | 0.12 |
ENST00000528301.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr9_-_21305312 | 0.11 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr17_+_7758374 | 0.11 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chrX_+_95939638 | 0.11 |
ENST00000373061.3
ENST00000373054.4 ENST00000355827.4 |
DIAPH2
|
diaphanous-related formin 2 |
| chr3_-_195310802 | 0.11 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chrX_-_119693745 | 0.11 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr3_+_44840679 | 0.11 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr10_-_14050522 | 0.10 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr17_+_61473104 | 0.10 |
ENST00000583016.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr17_-_48943706 | 0.10 |
ENST00000499247.2
|
TOB1
|
transducer of ERBB2, 1 |
| chr18_-_68004529 | 0.10 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr6_-_131299929 | 0.10 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr7_-_83824449 | 0.10 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr18_+_56532100 | 0.10 |
ENST00000588456.1
ENST00000589481.1 ENST00000591049.1 |
ZNF532
|
zinc finger protein 532 |
| chr1_-_31666767 | 0.10 |
ENST00000530145.1
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr5_+_68485433 | 0.10 |
ENST00000502689.1
|
CENPH
|
centromere protein H |
| chr5_-_111754948 | 0.10 |
ENST00000261486.5
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr10_+_91092241 | 0.10 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr4_-_99578776 | 0.09 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr12_-_122879969 | 0.09 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr8_+_16884740 | 0.09 |
ENST00000318063.5
|
MICU3
|
mitochondrial calcium uptake family, member 3 |
| chr7_-_35013217 | 0.09 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr2_-_114461655 | 0.09 |
ENST00000424612.1
|
AC017074.2
|
AC017074.2 |
| chr5_-_43515231 | 0.09 |
ENST00000306862.2
|
C5orf34
|
chromosome 5 open reading frame 34 |
| chr12_+_27398584 | 0.09 |
ENST00000543246.1
|
STK38L
|
serine/threonine kinase 38 like |
| chr15_+_59730348 | 0.09 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr14_+_78174414 | 0.09 |
ENST00000557342.1
ENST00000238688.5 ENST00000557623.1 ENST00000557431.1 ENST00000556831.1 ENST00000556375.1 ENST00000553981.1 |
SLIRP
|
SRA stem-loop interacting RNA binding protein |
| chr14_-_31926623 | 0.09 |
ENST00000356180.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr17_-_39623681 | 0.09 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr4_-_76944621 | 0.09 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr10_-_128210005 | 0.09 |
ENST00000284694.7
ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr12_+_16500037 | 0.09 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr10_+_79793518 | 0.08 |
ENST00000440692.1
ENST00000435275.1 ENST00000372360.3 ENST00000360830.4 |
RPS24
|
ribosomal protein S24 |
| chr7_+_104581389 | 0.08 |
ENST00000415513.1
ENST00000417026.1 |
RP11-325F22.2
|
RP11-325F22.2 |
| chr1_-_43282906 | 0.08 |
ENST00000372521.4
|
CCDC23
|
coiled-coil domain containing 23 |
| chr5_+_5422778 | 0.08 |
ENST00000296564.7
|
KIAA0947
|
KIAA0947 |
| chrX_-_130037162 | 0.08 |
ENST00000432489.1
|
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr1_+_150254936 | 0.08 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr2_+_103089756 | 0.08 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr12_-_88535747 | 0.08 |
ENST00000309041.7
|
CEP290
|
centrosomal protein 290kDa |
| chrX_+_95939711 | 0.08 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr4_+_187812086 | 0.08 |
ENST00000507644.2
|
RP11-11N5.1
|
RP11-11N5.1 |
| chr17_+_57297807 | 0.08 |
ENST00000284116.4
ENST00000581140.1 ENST00000581276.1 |
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr14_+_45464658 | 0.08 |
ENST00000555874.1
|
FAM179B
|
family with sequence similarity 179, member B |
| chr10_+_85933494 | 0.08 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr17_-_29151686 | 0.08 |
ENST00000544695.1
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr13_-_46425865 | 0.07 |
ENST00000400405.2
|
SIAH3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr11_-_3400442 | 0.07 |
ENST00000429541.2
ENST00000532539.1 |
ZNF195
|
zinc finger protein 195 |
| chr8_-_101730061 | 0.07 |
ENST00000519100.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr15_-_31283618 | 0.07 |
ENST00000563714.1
|
MTMR10
|
myotubularin related protein 10 |
| chr11_-_76155618 | 0.07 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr18_+_21033239 | 0.07 |
ENST00000581585.1
ENST00000577501.1 |
RIOK3
|
RIO kinase 3 |
| chr14_+_23938891 | 0.07 |
ENST00000408901.3
ENST00000397154.3 ENST00000555128.1 |
NGDN
|
neuroguidin, EIF4E binding protein |
| chr3_+_87276407 | 0.07 |
ENST00000471660.1
ENST00000263780.4 ENST00000494980.1 |
CHMP2B
|
charged multivesicular body protein 2B |
| chr16_+_15596123 | 0.07 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr7_-_99277610 | 0.07 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr7_+_16793160 | 0.07 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chrX_-_130037198 | 0.07 |
ENST00000370935.1
ENST00000338144.3 ENST00000394363.1 |
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr7_+_65338312 | 0.07 |
ENST00000434382.2
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chrX_+_70503526 | 0.07 |
ENST00000413858.1
ENST00000450092.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr11_+_101918153 | 0.07 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr19_-_18314836 | 0.07 |
ENST00000464076.3
ENST00000222256.4 |
RAB3A
|
RAB3A, member RAS oncogene family |
| chr1_-_197169672 | 0.07 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr10_-_115904361 | 0.07 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr3_+_197687071 | 0.07 |
ENST00000482695.1
ENST00000330198.4 ENST00000419117.1 ENST00000420910.2 ENST00000332636.5 |
LMLN
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr22_+_23248512 | 0.07 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr17_+_73663470 | 0.07 |
ENST00000583536.1
|
SAP30BP
|
SAP30 binding protein |
| chr2_+_109204909 | 0.07 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr5_+_176731572 | 0.07 |
ENST00000503853.1
|
PRELID1
|
PRELI domain containing 1 |
| chr6_-_137539651 | 0.07 |
ENST00000543628.1
|
IFNGR1
|
interferon gamma receptor 1 |
| chr2_-_121624973 | 0.07 |
ENST00000603720.1
|
RP11-297J22.1
|
RP11-297J22.1 |
| chrX_-_45629661 | 0.06 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr2_+_67624430 | 0.06 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr3_+_136649311 | 0.06 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr2_+_234104079 | 0.06 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr6_-_133035185 | 0.06 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr3_+_157827841 | 0.06 |
ENST00000295930.3
ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr19_+_37341752 | 0.06 |
ENST00000586933.1
ENST00000532141.1 ENST00000420450.1 ENST00000526123.1 |
ZNF345
|
zinc finger protein 345 |
| chr8_+_76452097 | 0.06 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr1_+_226250379 | 0.06 |
ENST00000366815.3
ENST00000366814.3 |
H3F3A
|
H3 histone, family 3A |
| chr5_+_75904918 | 0.06 |
ENST00000514001.1
ENST00000396234.3 ENST00000509074.1 |
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr15_+_79165112 | 0.06 |
ENST00000426013.2
|
MORF4L1
|
mortality factor 4 like 1 |
| chr11_+_71927807 | 0.06 |
ENST00000298223.6
ENST00000454954.2 ENST00000541003.1 ENST00000539412.1 ENST00000536778.1 ENST00000535625.1 ENST00000321324.7 |
FOLR2
|
folate receptor 2 (fetal) |
| chr19_+_8455077 | 0.06 |
ENST00000328024.6
|
RAB11B
|
RAB11B, member RAS oncogene family |
| chr13_-_30881621 | 0.06 |
ENST00000380615.3
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr15_+_33022885 | 0.06 |
ENST00000322805.4
|
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr3_-_196911002 | 0.06 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr4_-_159644507 | 0.06 |
ENST00000307720.3
|
PPID
|
peptidylprolyl isomerase D |
| chr22_+_32754139 | 0.06 |
ENST00000382088.3
|
RFPL3
|
ret finger protein-like 3 |
| chr1_-_33168336 | 0.06 |
ENST00000373484.3
|
SYNC
|
syncoilin, intermediate filament protein |
| chr14_+_89290965 | 0.06 |
ENST00000345383.5
ENST00000536576.1 ENST00000346301.4 ENST00000338104.6 ENST00000354441.6 ENST00000380656.2 ENST00000556651.1 ENST00000554686.1 |
TTC8
|
tetratricopeptide repeat domain 8 |
| chr12_+_32687221 | 0.06 |
ENST00000525053.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr7_-_27169801 | 0.06 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr21_+_35014829 | 0.06 |
ENST00000451686.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr2_+_143635067 | 0.06 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chrX_+_41583408 | 0.06 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr19_-_52598958 | 0.06 |
ENST00000594440.1
ENST00000426391.2 ENST00000389534.4 |
ZNF841
|
zinc finger protein 841 |
| chr9_+_21440440 | 0.06 |
ENST00000276927.1
|
IFNA1
|
interferon, alpha 1 |
| chr18_-_72265035 | 0.06 |
ENST00000585279.1
ENST00000580048.1 |
LINC00909
|
long intergenic non-protein coding RNA 909 |
| chr15_-_49338748 | 0.06 |
ENST00000559471.1
|
SECISBP2L
|
SECIS binding protein 2-like |
| chr2_+_128377550 | 0.06 |
ENST00000437387.1
ENST00000409090.1 |
MYO7B
|
myosin VIIB |
| chr2_-_204399976 | 0.06 |
ENST00000457812.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr9_-_88356789 | 0.06 |
ENST00000357081.3
ENST00000376081.4 ENST00000337006.4 ENST00000376109.3 |
AGTPBP1
|
ATP/GTP binding protein 1 |
| chr22_-_31324215 | 0.06 |
ENST00000429468.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr11_-_35441597 | 0.06 |
ENST00000395753.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr19_-_6375860 | 0.06 |
ENST00000245810.1
|
PSPN
|
persephin |
| chr4_-_74964904 | 0.06 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr5_-_148758839 | 0.06 |
ENST00000261796.3
|
IL17B
|
interleukin 17B |
| chr7_+_69064566 | 0.06 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr5_+_34915444 | 0.06 |
ENST00000336767.5
|
BRIX1
|
BRX1, biogenesis of ribosomes, homolog (S. cerevisiae) |
| chr15_+_45315302 | 0.05 |
ENST00000267814.9
|
SORD
|
sorbitol dehydrogenase |
| chr17_+_58018269 | 0.05 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr22_+_42017987 | 0.05 |
ENST00000405506.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr9_-_104198042 | 0.05 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate |
| chr2_-_225434538 | 0.05 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr11_-_3400330 | 0.05 |
ENST00000427810.2
ENST00000005082.9 ENST00000534569.1 ENST00000438262.2 ENST00000528796.1 ENST00000528410.1 ENST00000529678.1 ENST00000354599.6 ENST00000526601.1 ENST00000525502.1 ENST00000533036.1 ENST00000399602.4 |
ZNF195
|
zinc finger protein 195 |
| chr5_-_150138061 | 0.05 |
ENST00000521533.1
ENST00000424236.1 |
DCTN4
|
dynactin 4 (p62) |
| chr16_+_16434185 | 0.05 |
ENST00000524823.2
|
AC138969.4
|
Protein PKD1P1 |
| chr18_-_72264805 | 0.05 |
ENST00000577806.1
|
LINC00909
|
long intergenic non-protein coding RNA 909 |
| chr8_-_28347737 | 0.05 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr11_+_7559485 | 0.05 |
ENST00000527790.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr10_+_5488564 | 0.05 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr1_-_193075180 | 0.05 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr11_-_104905840 | 0.05 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr4_-_69536346 | 0.05 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr1_+_198189921 | 0.05 |
ENST00000391974.3
|
NEK7
|
NIMA-related kinase 7 |
| chr10_+_92631709 | 0.05 |
ENST00000413330.1
ENST00000277882.3 |
RPP30
|
ribonuclease P/MRP 30kDa subunit |
| chr14_+_77582905 | 0.05 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr3_+_157154578 | 0.05 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr6_+_18155560 | 0.05 |
ENST00000546309.2
ENST00000388870.2 ENST00000397244.1 |
KDM1B
|
lysine (K)-specific demethylase 1B |
| chr10_+_124320156 | 0.05 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr8_-_97247759 | 0.05 |
ENST00000518406.1
ENST00000523920.1 ENST00000287022.5 |
UQCRB
|
ubiquinol-cytochrome c reductase binding protein |
| chr2_+_138721850 | 0.05 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr11_-_10920714 | 0.05 |
ENST00000533941.1
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr1_-_144866711 | 0.05 |
ENST00000530130.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr10_-_24911746 | 0.05 |
ENST00000320481.6
|
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr7_+_102553430 | 0.05 |
ENST00000339431.4
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr20_-_48732472 | 0.05 |
ENST00000340309.3
ENST00000415862.2 ENST00000371677.3 ENST00000420027.2 |
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr2_+_136343820 | 0.05 |
ENST00000410054.1
|
R3HDM1
|
R3H domain containing 1 |
| chr5_-_94417314 | 0.05 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr4_-_76439483 | 0.05 |
ENST00000380840.2
ENST00000513257.1 ENST00000507014.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr2_-_204400013 | 0.05 |
ENST00000374489.2
ENST00000374488.2 ENST00000308091.4 ENST00000453034.1 ENST00000420371.1 |
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr11_+_59522837 | 0.05 |
ENST00000437946.2
|
STX3
|
syntaxin 3 |
| chr10_+_51572339 | 0.05 |
ENST00000344348.6
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr19_-_50528584 | 0.04 |
ENST00000594092.1
ENST00000443401.2 ENST00000594948.1 ENST00000377011.2 ENST00000593919.1 ENST00000601324.1 ENST00000316763.3 ENST00000601341.1 ENST00000600259.1 |
VRK3
|
vaccinia related kinase 3 |
| chr12_-_371994 | 0.04 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr4_+_86749045 | 0.04 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr10_-_96829246 | 0.04 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr2_+_219646462 | 0.04 |
ENST00000258415.4
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr15_+_79165151 | 0.04 |
ENST00000331268.5
|
MORF4L1
|
mortality factor 4 like 1 |
| chr2_+_109204743 | 0.04 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr11_-_35441524 | 0.04 |
ENST00000395750.1
ENST00000449068.1 |
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr2_-_130031335 | 0.04 |
ENST00000375987.3
|
AC079586.1
|
AC079586.1 |
| chr22_-_18111499 | 0.04 |
ENST00000413576.1
ENST00000399796.2 ENST00000399798.2 ENST00000253413.5 |
ATP6V1E1
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E1 |
| chr1_+_104615595 | 0.04 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr1_-_207095324 | 0.04 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr5_-_94417339 | 0.04 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr10_-_94301107 | 0.04 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr11_+_19138670 | 0.04 |
ENST00000446113.2
ENST00000399351.3 |
ZDHHC13
|
zinc finger, DHHC-type containing 13 |
| chr3_+_141103634 | 0.04 |
ENST00000507722.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr4_+_102711874 | 0.04 |
ENST00000508653.1
|
BANK1
|
B-cell scaffold protein with ankyrin repeats 1 |
| chr2_+_201170596 | 0.04 |
ENST00000439084.1
ENST00000409718.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr19_+_44645700 | 0.04 |
ENST00000592437.1
|
ZNF234
|
zinc finger protein 234 |
| chr7_-_83824169 | 0.04 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr3_+_57875738 | 0.04 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chr1_+_24646002 | 0.04 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr8_-_42358742 | 0.04 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr16_-_67427389 | 0.04 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr4_+_48018781 | 0.04 |
ENST00000295461.5
|
NIPAL1
|
NIPA-like domain containing 1 |
| chr12_+_107168342 | 0.04 |
ENST00000392837.4
|
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chrX_+_114827818 | 0.04 |
ENST00000420625.2
|
PLS3
|
plastin 3 |
| chr2_+_44396000 | 0.04 |
ENST00000409895.4
ENST00000409432.3 ENST00000282412.4 ENST00000378551.2 ENST00000345249.4 |
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
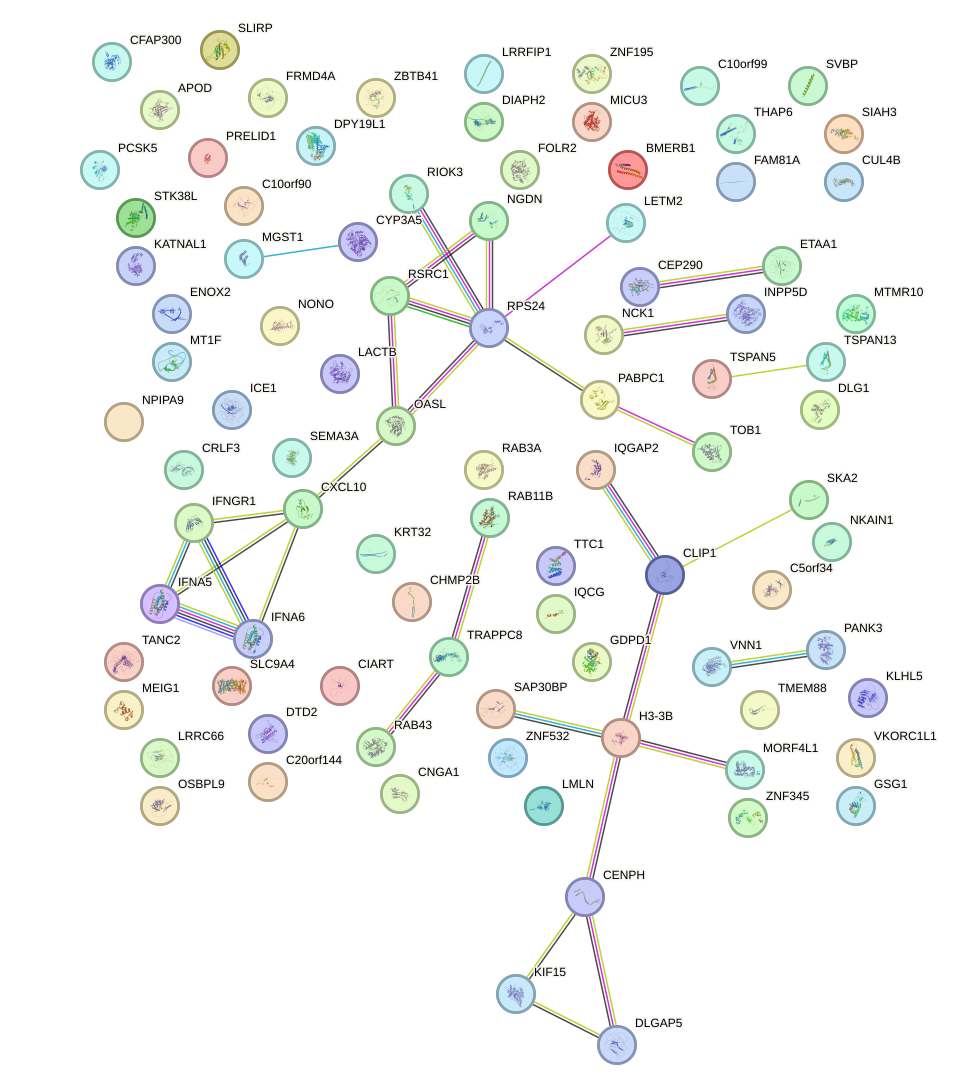
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.2 | GO:0001999 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) renin secretion into blood stream(GO:0002001) |
| 0.0 | 0.1 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0060623 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) |
| 0.0 | 0.1 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.0 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.0 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.0 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.0 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |