Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
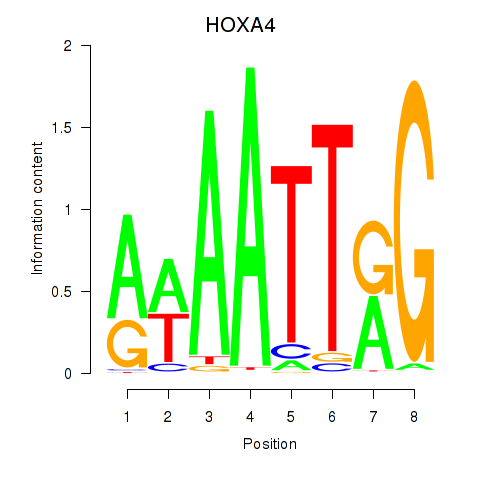
Results for HOXA4
Z-value: 0.84
Transcription factors associated with HOXA4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA4
|
ENSG00000197576.9 | homeobox A4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA4 | hg19_v2_chr7_-_27169801_27169844 | -0.64 | 3.6e-01 | Click! |
Activity profile of HOXA4 motif
Sorted Z-values of HOXA4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_66918558 | 0.71 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr5_-_56778635 | 0.67 |
ENST00000423391.1
|
ACTBL2
|
actin, beta-like 2 |
| chr9_-_3469181 | 0.55 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr6_-_26235206 | 0.46 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr10_-_92681033 | 0.40 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr3_-_64253655 | 0.35 |
ENST00000498162.1
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr11_+_128563652 | 0.34 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr19_+_45409011 | 0.28 |
ENST00000252486.4
ENST00000446996.1 ENST00000434152.1 |
APOE
|
apolipoprotein E |
| chr18_-_56985873 | 0.25 |
ENST00000299721.3
|
CPLX4
|
complexin 4 |
| chr12_+_72058130 | 0.24 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr20_+_57464200 | 0.21 |
ENST00000604005.1
|
GNAS
|
GNAS complex locus |
| chr14_-_36988882 | 0.21 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr2_-_190927447 | 0.21 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr11_+_112046190 | 0.20 |
ENST00000357685.5
ENST00000393032.2 ENST00000361053.4 |
BCO2
|
beta-carotene oxygenase 2 |
| chr6_-_31514333 | 0.20 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr19_+_12949251 | 0.19 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr6_+_63921351 | 0.18 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr18_-_56985776 | 0.18 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr14_-_36989336 | 0.18 |
ENST00000522719.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr17_-_67138015 | 0.17 |
ENST00000284425.2
ENST00000590645.1 |
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr21_+_17909594 | 0.17 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_-_2117600 | 0.17 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr7_-_130598059 | 0.16 |
ENST00000432045.2
|
MIR29B1
|
microRNA 29a |
| chr3_+_159557637 | 0.16 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr14_-_54423529 | 0.15 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr1_+_155658849 | 0.14 |
ENST00000368336.5
ENST00000343043.3 ENST00000421487.2 ENST00000535183.1 ENST00000465375.1 ENST00000470830.1 |
DAP3
|
death associated protein 3 |
| chr18_+_3247779 | 0.14 |
ENST00000578611.1
ENST00000583449.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr11_-_104480019 | 0.14 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr10_-_75415825 | 0.14 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr5_+_133562095 | 0.14 |
ENST00000602919.1
|
CTD-2410N18.3
|
CTD-2410N18.3 |
| chr17_-_48785216 | 0.13 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr6_-_116833500 | 0.13 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr14_+_101359265 | 0.13 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr1_+_28586006 | 0.13 |
ENST00000253063.3
|
SESN2
|
sestrin 2 |
| chr3_-_178984759 | 0.12 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr3_+_69134124 | 0.12 |
ENST00000478935.1
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr7_-_14026123 | 0.12 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr3_+_88188254 | 0.11 |
ENST00000309495.5
|
ZNF654
|
zinc finger protein 654 |
| chrX_-_55024967 | 0.11 |
ENST00000545676.1
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr7_-_29235063 | 0.10 |
ENST00000437527.1
ENST00000455544.1 |
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr7_-_14026063 | 0.10 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr12_+_56521840 | 0.10 |
ENST00000394048.5
|
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr1_-_12677714 | 0.10 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr2_+_39117010 | 0.10 |
ENST00000409978.1
|
ARHGEF33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr21_+_17442799 | 0.09 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_+_214161272 | 0.09 |
ENST00000498508.2
ENST00000366958.4 |
PROX1
|
prospero homeobox 1 |
| chr15_+_75080883 | 0.08 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr7_+_95115210 | 0.08 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr14_+_45605127 | 0.08 |
ENST00000556036.1
ENST00000267430.5 |
FANCM
|
Fanconi anemia, complementation group M |
| chr11_+_22688150 | 0.08 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr12_+_54447637 | 0.08 |
ENST00000609810.1
ENST00000430889.2 |
HOXC4
HOXC4
|
homeobox C4 Homeobox protein Hox-C4 |
| chr19_-_42806919 | 0.08 |
ENST00000595530.1
ENST00000538771.1 ENST00000601865.1 |
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chrX_-_19689106 | 0.07 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr6_+_31514622 | 0.07 |
ENST00000376146.4
|
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr15_+_90744533 | 0.07 |
ENST00000411539.2
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr12_+_56522001 | 0.07 |
ENST00000267113.4
ENST00000541590.1 |
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr21_+_17443434 | 0.07 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr12_+_50144381 | 0.07 |
ENST00000552370.1
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr18_-_53019208 | 0.07 |
ENST00000562607.1
|
TCF4
|
transcription factor 4 |
| chr7_-_14028488 | 0.07 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr18_-_21017817 | 0.06 |
ENST00000542162.1
ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241
|
transmembrane protein 241 |
| chr2_+_101437487 | 0.06 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr17_-_39538550 | 0.06 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr1_-_42384343 | 0.06 |
ENST00000372584.1
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr5_-_88179302 | 0.06 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr3_+_122103014 | 0.06 |
ENST00000232125.5
ENST00000477892.1 ENST00000469967.1 |
FAM162A
|
family with sequence similarity 162, member A |
| chr1_+_151739131 | 0.06 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr17_+_37894570 | 0.05 |
ENST00000394211.3
|
GRB7
|
growth factor receptor-bound protein 7 |
| chr14_+_38065052 | 0.05 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr22_+_26138108 | 0.05 |
ENST00000536101.1
ENST00000335473.7 ENST00000407587.2 |
MYO18B
|
myosin XVIIIB |
| chr7_-_29234802 | 0.05 |
ENST00000449801.1
ENST00000409850.1 |
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr5_+_49962772 | 0.05 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr5_+_66300446 | 0.05 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr14_+_45605157 | 0.05 |
ENST00000542564.2
|
FANCM
|
Fanconi anemia, complementation group M |
| chr7_-_75241096 | 0.05 |
ENST00000420909.1
|
HIP1
|
huntingtin interacting protein 1 |
| chr15_+_80351977 | 0.05 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr8_-_99954788 | 0.04 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr6_+_100054606 | 0.04 |
ENST00000369215.4
|
PRDM13
|
PR domain containing 13 |
| chr2_-_145278475 | 0.04 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr8_-_141774467 | 0.04 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr3_+_52448539 | 0.04 |
ENST00000461861.1
|
PHF7
|
PHD finger protein 7 |
| chr1_-_100643765 | 0.04 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr7_+_100273736 | 0.04 |
ENST00000412215.1
ENST00000393924.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chrX_-_119445306 | 0.04 |
ENST00000371369.4
ENST00000440464.1 ENST00000519908.1 |
TMEM255A
|
transmembrane protein 255A |
| chr21_+_17443521 | 0.04 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr16_+_57728701 | 0.04 |
ENST00000569375.1
ENST00000360716.3 ENST00000569167.1 ENST00000394337.4 ENST00000563126.1 ENST00000336825.8 |
CCDC135
|
coiled-coil domain containing 135 |
| chr14_+_37131058 | 0.03 |
ENST00000361487.6
|
PAX9
|
paired box 9 |
| chr3_+_69134080 | 0.03 |
ENST00000273258.3
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr20_-_45035223 | 0.03 |
ENST00000450812.1
ENST00000290246.6 ENST00000439931.2 ENST00000396391.1 |
ELMO2
|
engulfment and cell motility 2 |
| chr4_-_66536057 | 0.03 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chrX_-_119445263 | 0.03 |
ENST00000309720.5
|
TMEM255A
|
transmembrane protein 255A |
| chr15_+_51669444 | 0.03 |
ENST00000396399.2
|
GLDN
|
gliomedin |
| chr8_+_71383312 | 0.02 |
ENST00000520259.1
|
RP11-333A23.4
|
RP11-333A23.4 |
| chrX_+_37639264 | 0.02 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr2_+_54683419 | 0.02 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr19_-_51220176 | 0.02 |
ENST00000359082.3
ENST00000293441.1 |
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr4_-_109541539 | 0.02 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr14_+_59100774 | 0.02 |
ENST00000556859.1
ENST00000421793.1 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr12_+_12938541 | 0.02 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr8_+_77593474 | 0.02 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr13_-_46716969 | 0.02 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr1_+_245318279 | 0.02 |
ENST00000407071.2
|
KIF26B
|
kinesin family member 26B |
| chr7_+_32996997 | 0.02 |
ENST00000242209.4
ENST00000538336.1 ENST00000538443.1 |
FKBP9
|
FK506 binding protein 9, 63 kDa |
| chr3_-_129513259 | 0.01 |
ENST00000329333.5
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr15_+_80351910 | 0.01 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr10_+_11047259 | 0.01 |
ENST00000379261.4
ENST00000416382.2 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr7_+_144052381 | 0.01 |
ENST00000498580.1
ENST00000056217.5 |
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr12_+_53848505 | 0.01 |
ENST00000552819.1
ENST00000455667.3 |
PCBP2
|
poly(rC) binding protein 2 |
| chr1_+_170632250 | 0.01 |
ENST00000367760.3
|
PRRX1
|
paired related homeobox 1 |
| chr11_+_112832133 | 0.01 |
ENST00000524665.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr2_-_43453734 | 0.01 |
ENST00000282388.3
|
ZFP36L2
|
ZFP36 ring finger protein-like 2 |
| chr2_-_163099885 | 0.01 |
ENST00000443424.1
|
FAP
|
fibroblast activation protein, alpha |
| chr15_-_33447055 | 0.01 |
ENST00000559047.1
ENST00000561249.1 |
FMN1
|
formin 1 |
| chrX_+_37639302 | 0.01 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr4_-_152147579 | 0.01 |
ENST00000304527.4
ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19
|
SH3 domain containing 19 |
| chr21_+_22370717 | 0.01 |
ENST00000284894.7
|
NCAM2
|
neural cell adhesion molecule 2 |
| chr14_+_51026844 | 0.01 |
ENST00000554886.1
|
ATL1
|
atlastin GTPase 1 |
| chr8_+_77593448 | 0.01 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr6_+_50786414 | 0.01 |
ENST00000344788.3
ENST00000393655.3 ENST00000263046.4 |
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
| chr9_-_113761720 | 0.01 |
ENST00000541779.1
ENST00000374430.2 |
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr4_+_84457250 | 0.01 |
ENST00000395226.2
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr12_-_24103954 | 0.01 |
ENST00000441133.2
ENST00000545921.1 |
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr19_-_10047219 | 0.00 |
ENST00000264833.4
|
OLFM2
|
olfactomedin 2 |
| chr5_+_40909354 | 0.00 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr8_-_139926236 | 0.00 |
ENST00000303045.6
ENST00000435777.1 |
COL22A1
|
collagen, type XXII, alpha 1 |
| chr7_-_143892748 | 0.00 |
ENST00000378115.2
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35 |
| chr2_+_171571827 | 0.00 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr18_-_28742813 | 0.00 |
ENST00000257197.3
ENST00000257198.5 |
DSC1
|
desmocollin 1 |
| chr2_+_53994927 | 0.00 |
ENST00000295304.4
|
CHAC2
|
ChaC, cation transport regulator homolog 2 (E. coli) |
| chr14_+_39734482 | 0.00 |
ENST00000554392.1
ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5
|
CTAGE family, member 5 |
| chr2_-_163100045 | 0.00 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:2000646 | lipid transport involved in lipid storage(GO:0010877) regulation of Cdc42 protein signal transduction(GO:0032489) positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.2 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.2 | GO:0072099 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.2 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) negative regulation of amino acid transport(GO:0051956) |
| 0.0 | 0.0 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.1 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.1 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.3 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.4 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |