Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
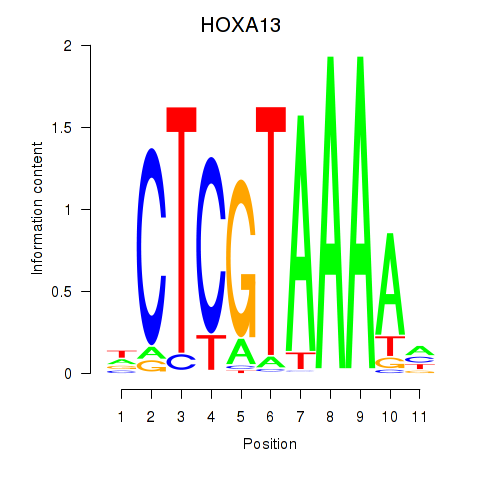
Results for HOXA13
Z-value: 0.55
Transcription factors associated with HOXA13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA13
|
ENSG00000106031.6 | homeobox A13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA13 | hg19_v2_chr7_-_27239703_27239725 | 0.30 | 7.0e-01 | Click! |
Activity profile of HOXA13 motif
Sorted Z-values of HOXA13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_15496722 | 0.39 |
ENST00000472534.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr19_+_35773242 | 0.24 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chr5_-_78808617 | 0.19 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr4_+_124571409 | 0.17 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr7_-_75401513 | 0.17 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr14_-_88282595 | 0.16 |
ENST00000554519.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chr3_+_113667354 | 0.15 |
ENST00000491556.1
|
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr11_-_46113756 | 0.14 |
ENST00000531959.1
|
PHF21A
|
PHD finger protein 21A |
| chr16_-_46865286 | 0.14 |
ENST00000285697.4
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr2_+_37571845 | 0.13 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr6_+_88117683 | 0.13 |
ENST00000369562.4
|
C6ORF165
|
UPF0704 protein C6orf165 |
| chr15_+_85523671 | 0.13 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chr3_+_57882061 | 0.13 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chr12_+_6309517 | 0.13 |
ENST00000382519.4
ENST00000009180.4 |
CD9
|
CD9 molecule |
| chr7_-_69062391 | 0.12 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr4_+_42399856 | 0.11 |
ENST00000319234.4
|
SHISA3
|
shisa family member 3 |
| chr5_+_158737824 | 0.11 |
ENST00000521472.1
|
AC008697.1
|
AC008697.1 |
| chrX_-_38186775 | 0.11 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr11_+_113185778 | 0.11 |
ENST00000524580.2
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr14_-_71067360 | 0.11 |
ENST00000554963.1
ENST00000430055.2 ENST00000440435.2 ENST00000256379.5 |
MED6
|
mediator complex subunit 6 |
| chr16_-_28192360 | 0.10 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr15_+_79166065 | 0.10 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr2_-_32390801 | 0.10 |
ENST00000608489.1
|
RP11-563N4.1
|
RP11-563N4.1 |
| chr17_-_8286484 | 0.10 |
ENST00000582556.1
ENST00000584164.1 ENST00000293842.5 ENST00000584343.1 ENST00000578812.1 ENST00000583011.1 |
RPL26
|
ribosomal protein L26 |
| chr2_+_37571717 | 0.10 |
ENST00000338415.3
ENST00000404976.1 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr8_-_73793975 | 0.10 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr14_-_71609965 | 0.10 |
ENST00000602957.1
|
RP6-91H8.3
|
RP6-91H8.3 |
| chr4_+_70894130 | 0.10 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr1_+_100818009 | 0.10 |
ENST00000370125.2
ENST00000361544.6 ENST00000370124.3 |
CDC14A
|
cell division cycle 14A |
| chr3_-_123680246 | 0.10 |
ENST00000488653.2
|
CCDC14
|
coiled-coil domain containing 14 |
| chr11_+_101785727 | 0.09 |
ENST00000263468.8
|
KIAA1377
|
KIAA1377 |
| chr3_+_46618727 | 0.09 |
ENST00000296145.5
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr1_-_31845914 | 0.09 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr11_+_94706804 | 0.09 |
ENST00000335080.5
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr9_+_137979506 | 0.09 |
ENST00000539529.1
ENST00000392991.4 ENST00000371793.3 |
OLFM1
|
olfactomedin 1 |
| chr4_+_54243917 | 0.08 |
ENST00000507166.1
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr8_+_94710789 | 0.08 |
ENST00000523475.1
|
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr4_+_128702969 | 0.08 |
ENST00000508776.1
ENST00000439123.2 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr19_-_6279932 | 0.08 |
ENST00000252674.7
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
| chr5_+_95385819 | 0.08 |
ENST00000507997.1
|
RP11-254I22.1
|
RP11-254I22.1 |
| chr19_+_39574945 | 0.08 |
ENST00000331256.5
|
PAPL
|
Iron/zinc purple acid phosphatase-like protein |
| chr3_-_156877997 | 0.08 |
ENST00000295926.3
|
CCNL1
|
cyclin L1 |
| chr14_-_92414294 | 0.08 |
ENST00000554468.1
|
FBLN5
|
fibulin 5 |
| chr14_+_97925151 | 0.08 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr3_+_72201910 | 0.08 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr10_-_13544945 | 0.08 |
ENST00000378605.3
ENST00000341083.3 |
BEND7
|
BEN domain containing 7 |
| chr1_+_19967014 | 0.07 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr17_-_56769382 | 0.07 |
ENST00000240361.8
ENST00000349033.5 ENST00000389934.3 |
TEX14
|
testis expressed 14 |
| chr5_-_40755987 | 0.07 |
ENST00000337702.4
|
TTC33
|
tetratricopeptide repeat domain 33 |
| chr5_-_150284532 | 0.07 |
ENST00000394226.2
ENST00000446148.2 ENST00000274599.5 ENST00000418587.2 |
ZNF300
|
zinc finger protein 300 |
| chr15_+_69373210 | 0.07 |
ENST00000435479.1
ENST00000559870.1 |
LINC00277
RP11-809H16.5
|
long intergenic non-protein coding RNA 277 RP11-809H16.5 |
| chr2_-_169104651 | 0.07 |
ENST00000355999.4
|
STK39
|
serine threonine kinase 39 |
| chr2_-_74007193 | 0.07 |
ENST00000377706.4
ENST00000443070.1 ENST00000272444.3 |
DUSP11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr3_-_123680047 | 0.07 |
ENST00000409697.3
|
CCDC14
|
coiled-coil domain containing 14 |
| chr3_-_160116995 | 0.07 |
ENST00000465537.1
ENST00000486856.1 ENST00000468218.1 ENST00000478370.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr7_+_141438393 | 0.07 |
ENST00000484178.1
ENST00000473783.1 ENST00000481508.1 |
SSBP1
|
single-stranded DNA binding protein 1, mitochondrial |
| chr12_+_97300995 | 0.07 |
ENST00000266742.4
ENST00000429527.2 ENST00000554226.1 ENST00000557478.1 ENST00000557092.1 ENST00000411739.2 ENST00000553609.1 |
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr11_+_111896090 | 0.07 |
ENST00000393051.1
|
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chr16_+_82068585 | 0.07 |
ENST00000563491.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr3_-_149093499 | 0.07 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr2_+_64681103 | 0.07 |
ENST00000464281.1
|
LGALSL
|
lectin, galactoside-binding-like |
| chr3_-_172241250 | 0.06 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr8_-_82608409 | 0.06 |
ENST00000518568.1
|
SLC10A5
|
solute carrier family 10, member 5 |
| chr1_-_207224307 | 0.06 |
ENST00000315927.4
|
YOD1
|
YOD1 deubiquitinase |
| chr11_-_4719072 | 0.06 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chr17_+_39382900 | 0.06 |
ENST00000377721.3
ENST00000455970.2 |
KRTAP9-2
|
keratin associated protein 9-2 |
| chr12_+_45609893 | 0.06 |
ENST00000320560.8
|
ANO6
|
anoctamin 6 |
| chr7_-_17598506 | 0.06 |
ENST00000451792.1
|
AC017060.1
|
AC017060.1 |
| chr4_-_170533723 | 0.06 |
ENST00000510533.1
ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1
|
NIMA-related kinase 1 |
| chr10_+_90750493 | 0.06 |
ENST00000357339.2
ENST00000355279.2 |
FAS
|
Fas cell surface death receptor |
| chr5_+_53686658 | 0.06 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chr3_+_50211240 | 0.06 |
ENST00000420831.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr14_+_56078695 | 0.06 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr4_-_17513604 | 0.06 |
ENST00000505710.1
|
QDPR
|
quinoid dihydropteridine reductase |
| chr4_-_82136114 | 0.06 |
ENST00000395578.1
ENST00000418486.2 |
PRKG2
|
protein kinase, cGMP-dependent, type II |
| chr8_+_26150628 | 0.06 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr12_-_8693539 | 0.06 |
ENST00000299663.3
|
CLEC4E
|
C-type lectin domain family 4, member E |
| chrX_+_13707235 | 0.06 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr7_-_151511911 | 0.06 |
ENST00000392801.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr1_-_17231271 | 0.06 |
ENST00000606899.1
|
RP11-108M9.6
|
RP11-108M9.6 |
| chr15_+_77713299 | 0.06 |
ENST00000559099.1
|
HMG20A
|
high mobility group 20A |
| chr1_-_150208498 | 0.06 |
ENST00000314136.8
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr5_-_150284351 | 0.06 |
ENST00000427179.1
|
ZNF300
|
zinc finger protein 300 |
| chr1_+_78511586 | 0.05 |
ENST00000370759.3
|
GIPC2
|
GIPC PDZ domain containing family, member 2 |
| chr1_+_222886694 | 0.05 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr12_-_111358372 | 0.05 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr9_-_21305312 | 0.05 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr20_+_37554955 | 0.05 |
ENST00000217429.4
|
FAM83D
|
family with sequence similarity 83, member D |
| chr3_-_170587815 | 0.05 |
ENST00000466674.1
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr5_+_169010638 | 0.05 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr19_-_6375860 | 0.05 |
ENST00000245810.1
|
PSPN
|
persephin |
| chr2_-_74007095 | 0.05 |
ENST00000452812.1
|
DUSP11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr9_-_69229650 | 0.05 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr17_-_57158523 | 0.05 |
ENST00000581468.1
|
TRIM37
|
tripartite motif containing 37 |
| chr12_+_75874580 | 0.05 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr17_-_40169659 | 0.05 |
ENST00000457167.4
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr4_+_54243862 | 0.05 |
ENST00000306932.6
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr9_-_134145880 | 0.05 |
ENST00000372269.3
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78, member A |
| chr10_-_30638090 | 0.05 |
ENST00000421701.1
ENST00000263063.4 |
MTPAP
|
mitochondrial poly(A) polymerase |
| chr10_+_103986085 | 0.05 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chrX_-_19688475 | 0.05 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr1_+_145525015 | 0.05 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr6_+_126307576 | 0.05 |
ENST00000334379.5
ENST00000450358.1 ENST00000368332.3 |
TRMT11
|
tRNA methyltransferase 11 homolog (S. cerevisiae) |
| chr2_+_109223595 | 0.05 |
ENST00000410093.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr4_+_72052964 | 0.05 |
ENST00000264485.5
ENST00000425175.1 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr11_+_29181503 | 0.05 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr14_-_50319758 | 0.05 |
ENST00000298310.5
|
NEMF
|
nuclear export mediator factor |
| chr12_+_59194154 | 0.05 |
ENST00000548969.1
|
RP11-362K2.2
|
Protein LOC100506869 |
| chr10_-_61513146 | 0.05 |
ENST00000430431.1
|
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chr16_+_28565230 | 0.05 |
ENST00000317058.3
|
CCDC101
|
coiled-coil domain containing 101 |
| chr12_-_4647606 | 0.05 |
ENST00000261250.3
ENST00000541014.1 ENST00000545746.1 ENST00000542080.1 |
C12orf4
|
chromosome 12 open reading frame 4 |
| chr1_+_171107241 | 0.05 |
ENST00000236166.3
|
FMO6P
|
flavin containing monooxygenase 6 pseudogene |
| chr2_+_33359473 | 0.05 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr11_+_111896320 | 0.05 |
ENST00000531306.1
ENST00000537636.1 |
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chr12_+_20963647 | 0.05 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr10_-_61513201 | 0.05 |
ENST00000414264.1
ENST00000594536.1 |
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chr5_-_89705537 | 0.05 |
ENST00000522864.1
ENST00000522083.1 ENST00000522565.1 ENST00000522842.1 ENST00000283122.3 |
CETN3
|
centrin, EF-hand protein, 3 |
| chr3_-_170587974 | 0.05 |
ENST00000463836.1
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr12_+_80838126 | 0.05 |
ENST00000266688.5
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q |
| chr12_+_45609862 | 0.05 |
ENST00000423947.3
ENST00000435642.1 |
ANO6
|
anoctamin 6 |
| chr11_+_122709200 | 0.05 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr3_+_99979828 | 0.04 |
ENST00000485687.1
ENST00000344949.5 ENST00000394144.4 |
TBC1D23
|
TBC1 domain family, member 23 |
| chr3_-_100712352 | 0.04 |
ENST00000471714.1
ENST00000284322.5 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr18_+_3262415 | 0.04 |
ENST00000581193.1
ENST00000400175.5 |
MYL12B
|
myosin, light chain 12B, regulatory |
| chr13_+_78315348 | 0.04 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr1_+_46769303 | 0.04 |
ENST00000311672.5
|
UQCRH
|
ubiquinol-cytochrome c reductase hinge protein |
| chr5_-_74532696 | 0.04 |
ENST00000506364.2
ENST00000274361.3 |
ANKRD31
|
ankyrin repeat domain 31 |
| chr10_+_92631709 | 0.04 |
ENST00000413330.1
ENST00000277882.3 |
RPP30
|
ribonuclease P/MRP 30kDa subunit |
| chr4_-_155533787 | 0.04 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr13_+_34392185 | 0.04 |
ENST00000380071.3
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr14_-_94789663 | 0.04 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr7_+_141438118 | 0.04 |
ENST00000265304.6
ENST00000498107.1 ENST00000467681.1 ENST00000465582.1 ENST00000463093.1 |
SSBP1
|
single-stranded DNA binding protein 1, mitochondrial |
| chr1_+_84609944 | 0.04 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr16_+_66914264 | 0.04 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chrX_+_117480036 | 0.04 |
ENST00000371822.5
ENST00000254029.3 ENST00000371825.3 |
WDR44
|
WD repeat domain 44 |
| chr17_-_39526052 | 0.04 |
ENST00000251646.3
|
KRT33B
|
keratin 33B |
| chr11_-_61849656 | 0.04 |
ENST00000524942.1
|
RP11-810P12.5
|
RP11-810P12.5 |
| chr6_+_71377567 | 0.04 |
ENST00000422334.2
|
SMAP1
|
small ArfGAP 1 |
| chr17_-_73150629 | 0.04 |
ENST00000356033.4
ENST00000405458.3 ENST00000409753.3 |
HN1
|
hematological and neurological expressed 1 |
| chrX_+_123097014 | 0.04 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr2_+_33359687 | 0.04 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr3_+_132316081 | 0.04 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr7_-_130597935 | 0.04 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr8_-_110346614 | 0.04 |
ENST00000239690.4
|
NUDCD1
|
NudC domain containing 1 |
| chr3_+_190333097 | 0.04 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr14_-_50319482 | 0.04 |
ENST00000546046.1
ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF
|
nuclear export mediator factor |
| chr4_+_106631966 | 0.04 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr5_+_95997885 | 0.04 |
ENST00000511097.2
|
CAST
|
calpastatin |
| chr9_-_98269481 | 0.04 |
ENST00000418258.1
ENST00000553011.1 ENST00000551845.1 |
PTCH1
|
patched 1 |
| chr6_-_128222103 | 0.04 |
ENST00000434358.1
ENST00000543064.1 ENST00000368248.2 |
THEMIS
|
thymocyte selection associated |
| chr6_+_71377473 | 0.04 |
ENST00000370452.3
ENST00000316999.5 ENST00000370455.3 |
SMAP1
|
small ArfGAP 1 |
| chr12_-_53207842 | 0.04 |
ENST00000458244.2
|
KRT4
|
keratin 4 |
| chr4_+_8321882 | 0.04 |
ENST00000509453.1
ENST00000503186.1 |
RP11-774O3.2
RP11-774O3.1
|
RP11-774O3.2 RP11-774O3.1 |
| chr16_+_82090028 | 0.04 |
ENST00000568090.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr16_+_20911174 | 0.04 |
ENST00000568663.1
|
LYRM1
|
LYR motif containing 1 |
| chr10_-_113943447 | 0.04 |
ENST00000369425.1
ENST00000348367.4 ENST00000423155.1 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr2_-_23747214 | 0.04 |
ENST00000430988.1
|
AC011239.1
|
Uncharacterized protein |
| chr7_-_27224795 | 0.04 |
ENST00000006015.3
|
HOXA11
|
homeobox A11 |
| chr22_+_45714361 | 0.04 |
ENST00000452238.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr4_-_83280767 | 0.04 |
ENST00000514671.1
|
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr15_+_71228826 | 0.04 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr8_-_100905363 | 0.04 |
ENST00000524245.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr2_+_33359646 | 0.04 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr18_+_9103957 | 0.03 |
ENST00000400033.1
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr15_-_55881135 | 0.03 |
ENST00000302000.6
|
PYGO1
|
pygopus family PHD finger 1 |
| chr4_+_54243798 | 0.03 |
ENST00000337488.6
ENST00000358575.5 ENST00000507922.1 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr18_+_60206744 | 0.03 |
ENST00000586834.1
|
ZCCHC2
|
zinc finger, CCHC domain containing 2 |
| chr12_+_20963632 | 0.03 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr2_+_204732487 | 0.03 |
ENST00000302823.3
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr3_+_111578027 | 0.03 |
ENST00000431670.2
ENST00000412622.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr10_-_51623203 | 0.03 |
ENST00000444743.1
ENST00000374065.3 ENST00000374064.3 ENST00000260867.4 |
TIMM23
|
translocase of inner mitochondrial membrane 23 homolog (yeast) |
| chr13_+_78315528 | 0.03 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr2_+_182756615 | 0.03 |
ENST00000431877.2
ENST00000320370.7 |
SSFA2
|
sperm specific antigen 2 |
| chr22_+_20193905 | 0.03 |
ENST00000609602.1
|
LINC00896
|
long intergenic non-protein coding RNA 896 |
| chr1_-_220220000 | 0.03 |
ENST00000366923.3
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr10_-_31288398 | 0.03 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr1_+_42928945 | 0.03 |
ENST00000428554.2
|
CCDC30
|
coiled-coil domain containing 30 |
| chr12_-_56367101 | 0.03 |
ENST00000549233.2
|
PMEL
|
premelanosome protein |
| chr16_+_75252878 | 0.03 |
ENST00000361017.4
|
CTRB1
|
chymotrypsinogen B1 |
| chr3_+_100428188 | 0.03 |
ENST00000418917.2
ENST00000490574.1 |
TFG
|
TRK-fused gene |
| chr7_+_129015671 | 0.03 |
ENST00000466993.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr18_+_3262954 | 0.03 |
ENST00000584539.1
|
MYL12B
|
myosin, light chain 12B, regulatory |
| chr3_+_57875738 | 0.03 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chr5_+_159895275 | 0.03 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr13_+_78315295 | 0.03 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr12_-_12491608 | 0.03 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr1_+_200842083 | 0.03 |
ENST00000304244.2
|
GPR25
|
G protein-coupled receptor 25 |
| chr16_-_18812746 | 0.03 |
ENST00000546206.2
ENST00000562819.1 ENST00000562234.2 ENST00000304414.7 ENST00000567078.2 |
ARL6IP1
RP11-1035H13.3
|
ADP-ribosylation factor-like 6 interacting protein 1 Uncharacterized protein |
| chr1_+_13910194 | 0.03 |
ENST00000376057.4
ENST00000510906.1 |
PDPN
|
podoplanin |
| chr3_-_118753716 | 0.03 |
ENST00000393775.2
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr3_-_156878540 | 0.03 |
ENST00000461804.1
|
CCNL1
|
cyclin L1 |
| chr6_+_121756809 | 0.03 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chr2_+_32390925 | 0.03 |
ENST00000440718.1
ENST00000379343.2 ENST00000282587.5 ENST00000435660.1 ENST00000538303.1 ENST00000357055.3 ENST00000406369.1 |
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr7_-_29235063 | 0.03 |
ENST00000437527.1
ENST00000455544.1 |
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr2_-_86850949 | 0.03 |
ENST00000237455.4
|
RNF103
|
ring finger protein 103 |
| chr3_+_113251143 | 0.03 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr3_+_4535025 | 0.03 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr4_-_69083720 | 0.03 |
ENST00000432593.3
|
TMPRSS11BNL
|
TMPRSS11B N-terminal like |
| chr1_-_46642154 | 0.03 |
ENST00000540385.1
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr12_+_121131970 | 0.03 |
ENST00000535656.1
|
MLEC
|
malectin |
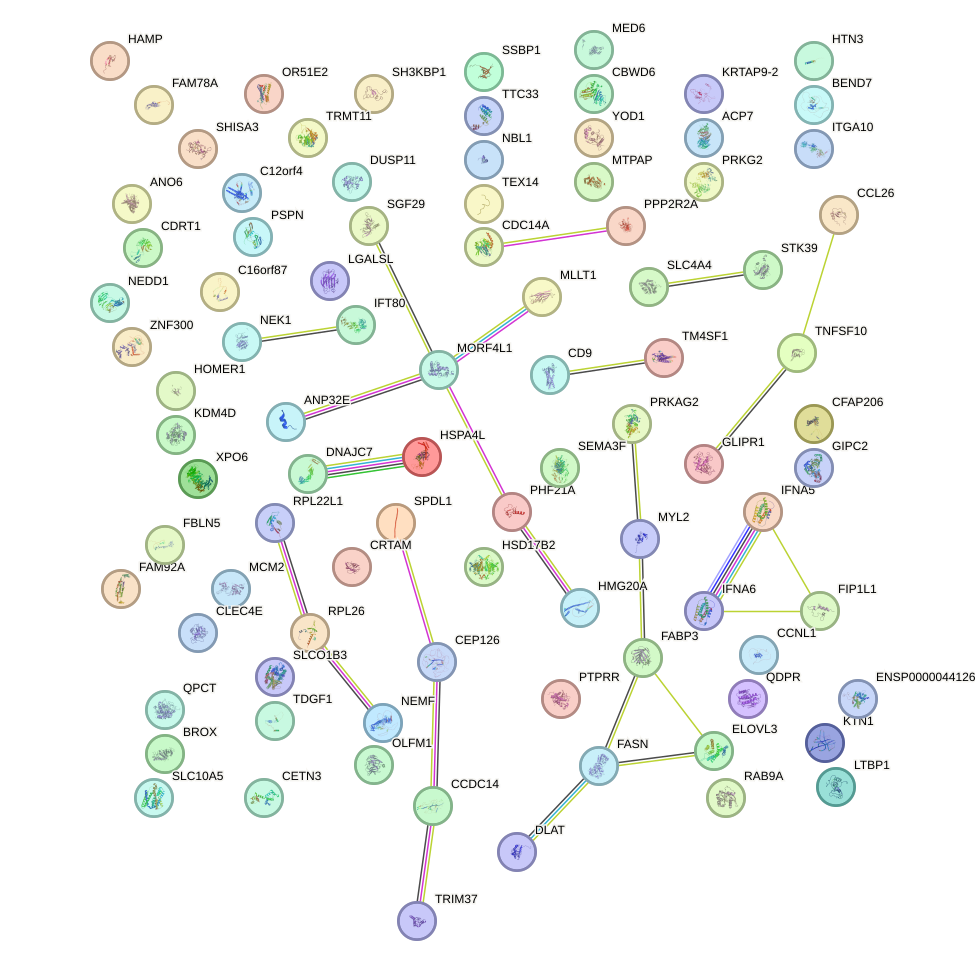
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903413 | cellular response to bile acid(GO:1903413) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.1 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.1 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.1 | GO:1902231 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.0 | GO:1904585 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.0 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.1 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.1 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.0 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |