Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
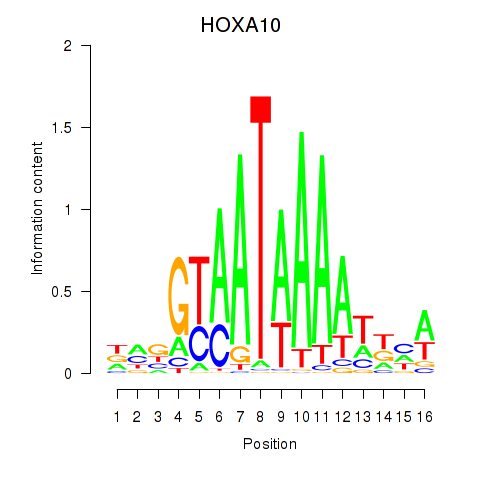
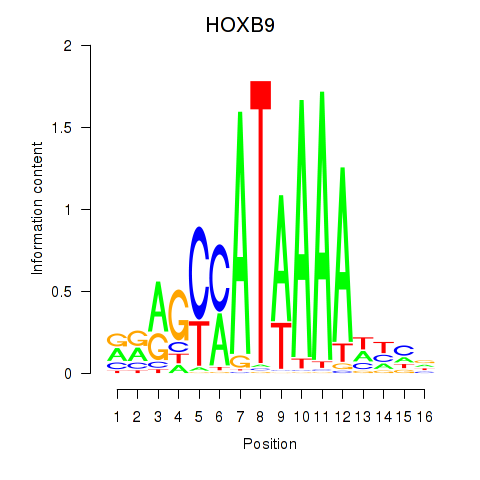
Results for HOXA10_HOXB9
Z-value: 0.20
Transcription factors associated with HOXA10_HOXB9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA10
|
ENSG00000253293.3 | homeobox A10 |
|
HOXB9
|
ENSG00000170689.8 | homeobox B9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA10 | hg19_v2_chr7_-_27219849_27219880 | 0.93 | 7.3e-02 | Click! |
| HOXB9 | hg19_v2_chr17_-_46703826_46703845 | 0.72 | 2.8e-01 | Click! |
Activity profile of HOXA10_HOXB9 motif
Sorted Z-values of HOXA10_HOXB9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_61151306 | 0.17 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr3_+_164924716 | 0.10 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr15_+_101417919 | 0.09 |
ENST00000561338.1
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr4_+_169633310 | 0.09 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr5_-_159766528 | 0.08 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr19_-_19626838 | 0.07 |
ENST00000360913.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr7_+_138915102 | 0.07 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr8_-_25281747 | 0.07 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr1_-_160231451 | 0.07 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr10_-_94301107 | 0.06 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr5_-_126409159 | 0.06 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr1_+_154401791 | 0.06 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr3_-_172241250 | 0.05 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr2_+_20101786 | 0.05 |
ENST00000607190.1
|
RP11-79O8.1
|
RP11-79O8.1 |
| chr5_+_135496675 | 0.05 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr22_-_38240316 | 0.05 |
ENST00000411961.2
ENST00000434930.1 |
ANKRD54
|
ankyrin repeat domain 54 |
| chr9_+_109685630 | 0.05 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr2_-_216878305 | 0.05 |
ENST00000263268.6
|
MREG
|
melanoregulin |
| chr9_-_104145795 | 0.05 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr14_+_35514323 | 0.05 |
ENST00000555211.1
|
FAM177A1
|
family with sequence similarity 177, member A1 |
| chr4_+_155484103 | 0.04 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr4_-_140222358 | 0.04 |
ENST00000505036.1
ENST00000544855.1 ENST00000539002.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr1_+_117963209 | 0.04 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr17_-_38821373 | 0.04 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr1_+_32379174 | 0.04 |
ENST00000391369.1
|
AL136115.1
|
HCG2032337; PRO1848; Uncharacterized protein |
| chr12_+_58166726 | 0.04 |
ENST00000546504.1
|
RP11-571M6.15
|
Uncharacterized protein |
| chrX_-_54070388 | 0.04 |
ENST00000415025.1
|
PHF8
|
PHD finger protein 8 |
| chr19_+_21106081 | 0.04 |
ENST00000300540.3
ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85
|
zinc finger protein 85 |
| chrX_-_54069253 | 0.04 |
ENST00000425862.1
ENST00000433120.1 |
PHF8
|
PHD finger protein 8 |
| chr12_+_57810198 | 0.03 |
ENST00000598001.1
|
AC126614.1
|
HCG1818482; Uncharacterized protein |
| chr20_+_42984330 | 0.03 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr9_+_2029019 | 0.03 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr11_-_104916034 | 0.03 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr1_-_242162375 | 0.03 |
ENST00000357246.3
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma |
| chr18_+_3466248 | 0.03 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr19_-_3500635 | 0.03 |
ENST00000250937.3
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase |
| chr7_-_50633078 | 0.03 |
ENST00000444124.2
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr12_+_64173583 | 0.03 |
ENST00000261234.6
|
TMEM5
|
transmembrane protein 5 |
| chr8_+_21906433 | 0.03 |
ENST00000522148.1
|
DMTN
|
dematin actin binding protein |
| chr15_+_67418047 | 0.03 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr4_+_155484155 | 0.03 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr8_-_95220775 | 0.03 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr7_-_41742697 | 0.03 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr6_+_80816342 | 0.03 |
ENST00000369760.4
ENST00000356489.5 ENST00000320393.6 |
BCKDHB
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr11_+_113185778 | 0.03 |
ENST00000524580.2
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr3_+_107364769 | 0.03 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr6_+_131958436 | 0.03 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr5_+_140514782 | 0.03 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr12_-_76879852 | 0.03 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr9_-_20382446 | 0.03 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr16_-_25122785 | 0.03 |
ENST00000563962.1
ENST00000569920.1 |
RP11-449H11.1
|
RP11-449H11.1 |
| chr11_+_63606373 | 0.02 |
ENST00000402010.2
ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr3_+_138340049 | 0.02 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr6_-_52859046 | 0.02 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr20_-_36152914 | 0.02 |
ENST00000397131.1
|
BLCAP
|
bladder cancer associated protein |
| chr7_+_128784712 | 0.02 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr1_-_161207986 | 0.02 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr4_-_89951028 | 0.02 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr4_-_129491686 | 0.02 |
ENST00000514265.1
|
RP11-184M15.1
|
RP11-184M15.1 |
| chr11_-_65359947 | 0.02 |
ENST00000597463.1
|
AP001362.1
|
Uncharacterized protein |
| chr19_-_19626351 | 0.02 |
ENST00000585580.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr2_-_176046391 | 0.02 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr14_+_31046959 | 0.02 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr5_-_58295712 | 0.02 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr6_+_143447322 | 0.02 |
ENST00000458219.1
|
AIG1
|
androgen-induced 1 |
| chr7_-_81635106 | 0.02 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr17_+_48823975 | 0.02 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr8_+_91233711 | 0.02 |
ENST00000523283.1
ENST00000517400.1 |
LINC00534
|
long intergenic non-protein coding RNA 534 |
| chr14_-_25479811 | 0.02 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr3_+_89156799 | 0.02 |
ENST00000452448.2
ENST00000494014.1 |
EPHA3
|
EPH receptor A3 |
| chr14_+_79745682 | 0.02 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr9_-_95640218 | 0.02 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr1_-_161208013 | 0.02 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr3_+_138340067 | 0.02 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr15_-_98417780 | 0.02 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chrX_+_55744228 | 0.02 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr1_+_12524965 | 0.02 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr9_-_133814455 | 0.02 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr3_+_185046676 | 0.02 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr2_+_138721850 | 0.02 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr4_+_146539415 | 0.02 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr1_-_230991747 | 0.02 |
ENST00000523410.1
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr12_+_75728419 | 0.02 |
ENST00000378695.4
ENST00000312442.2 |
GLIPR1L1
|
GLI pathogenesis-related 1 like 1 |
| chr11_+_47279155 | 0.02 |
ENST00000444396.1
ENST00000457932.1 ENST00000412937.1 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr7_+_57509877 | 0.02 |
ENST00000420713.1
|
ZNF716
|
zinc finger protein 716 |
| chr2_+_161993412 | 0.02 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr5_+_98109322 | 0.02 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr1_-_161207875 | 0.02 |
ENST00000512372.1
ENST00000437437.2 ENST00000442691.2 ENST00000412844.2 ENST00000428574.2 ENST00000505005.1 ENST00000508740.1 ENST00000508387.1 ENST00000504010.1 ENST00000511676.1 ENST00000502985.1 ENST00000367981.3 ENST00000515621.1 ENST00000511944.1 ENST00000511748.1 ENST00000367984.4 ENST00000367985.3 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr14_+_53173910 | 0.02 |
ENST00000606149.1
ENST00000555339.1 ENST00000556813.1 |
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr3_+_149191723 | 0.02 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr17_+_57807062 | 0.02 |
ENST00000587259.1
|
VMP1
|
vacuole membrane protein 1 |
| chr15_-_59500973 | 0.02 |
ENST00000560749.1
|
MYO1E
|
myosin IE |
| chr16_-_75498553 | 0.02 |
ENST00000569276.1
ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A
RP11-77K12.1
|
transmembrane protein 170A Uncharacterized protein |
| chr10_+_24755416 | 0.02 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr9_+_70856397 | 0.02 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr14_-_51027838 | 0.01 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr5_-_135290651 | 0.01 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr4_+_95972822 | 0.01 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr11_-_104827425 | 0.01 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr11_-_104972158 | 0.01 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr12_-_94673956 | 0.01 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr3_+_157828152 | 0.01 |
ENST00000476899.1
|
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr12_+_4130143 | 0.01 |
ENST00000543206.1
|
RP11-320N7.2
|
RP11-320N7.2 |
| chr9_+_124413873 | 0.01 |
ENST00000408936.3
|
DAB2IP
|
DAB2 interacting protein |
| chr5_-_111312622 | 0.01 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr20_+_10015678 | 0.01 |
ENST00000378392.1
ENST00000378380.3 |
ANKEF1
|
ankyrin repeat and EF-hand domain containing 1 |
| chr8_-_143859197 | 0.01 |
ENST00000395192.2
|
LYNX1
|
Ly6/neurotoxin 1 |
| chr12_-_118628350 | 0.01 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr2_-_231989808 | 0.01 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr21_+_35736302 | 0.01 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr10_+_91152303 | 0.01 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr11_-_46113756 | 0.01 |
ENST00000531959.1
|
PHF21A
|
PHD finger protein 21A |
| chr19_+_19626531 | 0.01 |
ENST00000507754.4
|
NDUFA13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr2_-_32390801 | 0.01 |
ENST00000608489.1
|
RP11-563N4.1
|
RP11-563N4.1 |
| chr1_-_23521222 | 0.01 |
ENST00000374619.1
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr1_-_155880672 | 0.01 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr5_+_158737824 | 0.01 |
ENST00000521472.1
|
AC008697.1
|
AC008697.1 |
| chr17_+_61086917 | 0.01 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr12_+_32638897 | 0.01 |
ENST00000531134.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr3_+_125687987 | 0.01 |
ENST00000514116.1
ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chrX_+_70503526 | 0.01 |
ENST00000413858.1
ENST00000450092.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr19_-_53636125 | 0.01 |
ENST00000601493.1
ENST00000599261.1 ENST00000597503.1 ENST00000500065.4 ENST00000243643.4 ENST00000594011.1 ENST00000455735.2 ENST00000595193.1 ENST00000448501.1 ENST00000421033.1 ENST00000440291.1 ENST00000595813.1 ENST00000600574.1 ENST00000596051.1 ENST00000601110.1 |
ZNF415
|
zinc finger protein 415 |
| chr8_-_8318847 | 0.01 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr2_+_220143989 | 0.01 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr5_-_159846399 | 0.01 |
ENST00000297151.4
|
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr22_-_23922448 | 0.01 |
ENST00000438703.1
ENST00000330377.2 |
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr8_+_91233750 | 0.01 |
ENST00000523406.1
|
LINC00534
|
long intergenic non-protein coding RNA 534 |
| chr14_-_38028689 | 0.01 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr18_-_52989217 | 0.01 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr10_-_73497581 | 0.01 |
ENST00000398786.2
|
C10orf105
|
chromosome 10 open reading frame 105 |
| chrX_-_133792480 | 0.01 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr13_+_24844979 | 0.01 |
ENST00000454083.1
|
SPATA13
|
spermatogenesis associated 13 |
| chr13_-_47471155 | 0.01 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr12_-_54071181 | 0.01 |
ENST00000338662.5
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr6_+_25279759 | 0.01 |
ENST00000377969.3
|
LRRC16A
|
leucine rich repeat containing 16A |
| chr21_+_34398153 | 0.01 |
ENST00000382357.3
ENST00000430860.1 ENST00000333337.3 |
OLIG2
|
oligodendrocyte lineage transcription factor 2 |
| chr4_-_104021009 | 0.01 |
ENST00000509245.1
ENST00000296424.4 |
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr12_-_12491608 | 0.01 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr3_-_196911002 | 0.01 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr3_-_150690786 | 0.01 |
ENST00000327047.1
|
CLRN1
|
clarin 1 |
| chr7_+_79763271 | 0.01 |
ENST00000442586.1
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr2_+_102927962 | 0.01 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr17_-_38928414 | 0.01 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr7_-_122840015 | 0.01 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr1_-_226926864 | 0.01 |
ENST00000429204.1
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr10_+_133747955 | 0.01 |
ENST00000455566.1
|
PPP2R2D
|
protein phosphatase 2, regulatory subunit B, delta |
| chr1_+_43613612 | 0.01 |
ENST00000335282.4
|
FAM183A
|
family with sequence similarity 183, member A |
| chr3_-_9921934 | 0.01 |
ENST00000423850.1
|
CIDEC
|
cell death-inducing DFFA-like effector c |
| chr11_-_62437486 | 0.01 |
ENST00000528115.1
|
C11orf48
|
chromosome 11 open reading frame 48 |
| chr14_+_73706308 | 0.01 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr2_+_197577841 | 0.01 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chr9_-_69262509 | 0.01 |
ENST00000377449.1
ENST00000382399.4 ENST00000377439.1 ENST00000377441.1 ENST00000377457.5 |
CBWD6
|
COBW domain containing 6 |
| chr11_-_104905840 | 0.01 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chrX_-_122756660 | 0.01 |
ENST00000441692.1
|
THOC2
|
THO complex 2 |
| chr6_+_158733692 | 0.01 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr7_-_77427676 | 0.01 |
ENST00000257663.3
|
TMEM60
|
transmembrane protein 60 |
| chrX_-_117119243 | 0.01 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr13_-_46626847 | 0.01 |
ENST00000242848.4
ENST00000282007.3 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr13_+_24844857 | 0.01 |
ENST00000409126.1
ENST00000343003.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr9_+_67977438 | 0.01 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr19_-_48753064 | 0.01 |
ENST00000520153.1
ENST00000357778.5 ENST00000520015.1 |
CARD8
|
caspase recruitment domain family, member 8 |
| chr2_+_208423891 | 0.01 |
ENST00000448277.1
ENST00000457101.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr16_-_15180257 | 0.01 |
ENST00000540462.1
|
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr22_-_22337146 | 0.01 |
ENST00000398793.2
|
TOP3B
|
topoisomerase (DNA) III beta |
| chr11_+_10471836 | 0.01 |
ENST00000444303.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr22_-_43010928 | 0.01 |
ENST00000348657.2
ENST00000252115.5 |
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr20_-_50384864 | 0.01 |
ENST00000311637.5
ENST00000402822.1 |
ATP9A
|
ATPase, class II, type 9A |
| chr1_+_53480598 | 0.01 |
ENST00000430330.2
ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr10_+_78078088 | 0.01 |
ENST00000496424.2
|
C10orf11
|
chromosome 10 open reading frame 11 |
| chr15_-_66797172 | 0.01 |
ENST00000569438.1
ENST00000569696.1 ENST00000307961.6 |
RPL4
|
ribosomal protein L4 |
| chr9_+_96846740 | 0.01 |
ENST00000288976.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr9_+_134065506 | 0.01 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr15_-_101817492 | 0.01 |
ENST00000528346.1
ENST00000531964.1 |
VIMP
|
VCP-interacting membrane protein |
| chr19_+_50016610 | 0.01 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr2_+_38177620 | 0.01 |
ENST00000402091.3
|
RMDN2
|
regulator of microtubule dynamics 2 |
| chr4_+_71017791 | 0.01 |
ENST00000502294.1
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr17_-_73663245 | 0.01 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chr17_-_73663168 | 0.01 |
ENST00000578201.1
ENST00000423245.2 |
RECQL5
|
RecQ protein-like 5 |
| chr1_-_17766198 | 0.01 |
ENST00000375436.4
|
RCC2
|
regulator of chromosome condensation 2 |
| chr14_+_79745746 | 0.01 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr1_+_52682052 | 0.01 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr14_+_53173890 | 0.01 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr15_+_52155001 | 0.01 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr18_+_43307294 | 0.01 |
ENST00000590246.1
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr18_-_10855434 | 0.01 |
ENST00000579112.1
|
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr8_+_146024261 | 0.01 |
ENST00000359971.3
ENST00000528012.1 |
ZNF517
|
zinc finger protein 517 |
| chr7_-_7782204 | 0.01 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr7_+_56131917 | 0.01 |
ENST00000434526.2
ENST00000275607.9 ENST00000395435.2 ENST00000413952.2 ENST00000342190.6 ENST00000437307.2 ENST00000413756.1 ENST00000451338.1 |
SUMF2
|
sulfatase modifying factor 2 |
| chr5_-_16738451 | 0.01 |
ENST00000274203.9
ENST00000515803.1 |
MYO10
|
myosin X |
| chr4_+_113568207 | 0.01 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr1_-_23520755 | 0.01 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr16_+_71560154 | 0.01 |
ENST00000539698.3
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr3_-_180707306 | 0.01 |
ENST00000479269.1
|
DNAJC19
|
DnaJ (Hsp40) homolog, subfamily C, member 19 |
| chr12_+_1800179 | 0.01 |
ENST00000357103.4
|
ADIPOR2
|
adiponectin receptor 2 |
| chr8_+_77593448 | 0.01 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr13_-_79979919 | 0.01 |
ENST00000267229.7
|
RBM26
|
RNA binding motif protein 26 |
| chr15_+_57891609 | 0.01 |
ENST00000569089.1
|
MYZAP
|
myocardial zonula adherens protein |
| chr8_+_145215928 | 0.01 |
ENST00000528919.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr6_+_148663729 | 0.01 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr1_-_161207953 | 0.01 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA10_HOXB9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.0 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.0 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |