Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
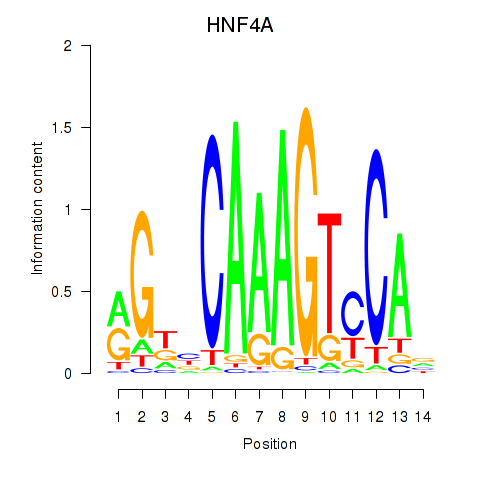
Results for HNF4A
Z-value: 1.03
Transcription factors associated with HNF4A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF4A
|
ENSG00000101076.12 | hepatocyte nuclear factor 4 alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HNF4A | hg19_v2_chr20_+_43029911_43029941 | 0.99 | 1.1e-02 | Click! |
Activity profile of HNF4A motif
Sorted Z-values of HNF4A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_73606766 | 1.12 |
ENST00000578462.1
|
MYO15B
|
myosin XVB pseudogene |
| chr19_+_42773371 | 1.09 |
ENST00000571942.2
|
CIC
|
capicua transcriptional repressor |
| chr16_-_11692320 | 0.64 |
ENST00000571627.1
|
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr1_+_145727681 | 0.57 |
ENST00000417171.1
ENST00000451928.2 |
PDZK1
|
PDZ domain containing 1 |
| chr3_-_53878644 | 0.53 |
ENST00000481668.1
ENST00000467802.1 |
CHDH
|
choline dehydrogenase |
| chr2_-_74648702 | 0.51 |
ENST00000518863.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr11_-_45939565 | 0.48 |
ENST00000525192.1
ENST00000378750.5 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr3_-_120400960 | 0.46 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr11_-_65381643 | 0.46 |
ENST00000309100.3
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr19_-_49339080 | 0.45 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr4_-_120243545 | 0.43 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr2_-_74645669 | 0.39 |
ENST00000518401.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr11_-_45939374 | 0.39 |
ENST00000533151.1
ENST00000241041.3 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr17_+_4692230 | 0.37 |
ENST00000331264.7
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr16_-_57514277 | 0.36 |
ENST00000562008.1
ENST00000567214.1 |
DOK4
|
docking protein 4 |
| chr1_-_177939041 | 0.36 |
ENST00000308284.6
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr12_+_56114151 | 0.32 |
ENST00000547072.1
ENST00000552930.1 ENST00000257895.5 |
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr11_+_64073699 | 0.32 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr2_+_201987200 | 0.31 |
ENST00000425030.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr1_+_43855545 | 0.31 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr19_-_10687907 | 0.31 |
ENST00000589348.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr22_+_27068704 | 0.31 |
ENST00000444388.1
ENST00000450963.1 ENST00000449017.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr12_+_121163602 | 0.30 |
ENST00000411593.2
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr14_+_23790655 | 0.28 |
ENST00000397276.2
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr9_+_139839686 | 0.28 |
ENST00000371634.2
|
C8G
|
complement component 8, gamma polypeptide |
| chr1_-_161207875 | 0.27 |
ENST00000512372.1
ENST00000437437.2 ENST00000442691.2 ENST00000412844.2 ENST00000428574.2 ENST00000505005.1 ENST00000508740.1 ENST00000508387.1 ENST00000504010.1 ENST00000511676.1 ENST00000502985.1 ENST00000367981.3 ENST00000515621.1 ENST00000511944.1 ENST00000511748.1 ENST00000367984.4 ENST00000367985.3 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr19_-_14201776 | 0.26 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr12_+_121163538 | 0.26 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr19_+_10196981 | 0.26 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr1_+_45274154 | 0.26 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr16_+_81272287 | 0.25 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr2_-_219134343 | 0.25 |
ENST00000447885.1
ENST00000420660.1 |
AAMP
|
angio-associated, migratory cell protein |
| chr17_+_48423453 | 0.24 |
ENST00000017003.2
ENST00000509778.1 ENST00000507602.1 |
XYLT2
|
xylosyltransferase II |
| chr2_-_228244013 | 0.23 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr2_+_10442993 | 0.22 |
ENST00000423674.1
ENST00000307845.3 |
HPCAL1
|
hippocalcin-like 1 |
| chr7_+_192969 | 0.21 |
ENST00000313766.5
|
FAM20C
|
family with sequence similarity 20, member C |
| chrX_+_66764375 | 0.21 |
ENST00000374690.3
|
AR
|
androgen receptor |
| chr1_-_26197744 | 0.21 |
ENST00000374296.3
|
PAQR7
|
progestin and adipoQ receptor family member VII |
| chr6_+_30881982 | 0.20 |
ENST00000321897.5
ENST00000416670.2 ENST00000542001.1 ENST00000428017.1 |
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr20_-_36156125 | 0.20 |
ENST00000397135.1
ENST00000397137.1 |
BLCAP
|
bladder cancer associated protein |
| chr12_-_109219937 | 0.20 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr12_+_56075330 | 0.20 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr8_+_22436635 | 0.20 |
ENST00000452226.1
ENST00000397760.4 ENST00000339162.7 ENST00000397761.2 |
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr19_-_10687948 | 0.20 |
ENST00000592285.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr11_-_61659006 | 0.19 |
ENST00000278829.2
|
FADS3
|
fatty acid desaturase 3 |
| chr6_+_30882108 | 0.19 |
ENST00000541562.1
ENST00000421263.1 |
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr5_-_42811986 | 0.19 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr19_+_18496957 | 0.19 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr12_+_121416489 | 0.18 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr9_+_137967395 | 0.18 |
ENST00000371801.4
|
OLFM1
|
olfactomedin 1 |
| chr1_+_23695680 | 0.18 |
ENST00000454117.1
ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213
|
chromosome 1 open reading frame 213 |
| chr12_+_121416340 | 0.18 |
ENST00000257555.6
ENST00000400024.2 |
HNF1A
|
HNF1 homeobox A |
| chr19_-_48867171 | 0.17 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr6_+_43028182 | 0.17 |
ENST00000394058.1
|
KLC4
|
kinesin light chain 4 |
| chr17_-_27503770 | 0.17 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr1_+_113217309 | 0.17 |
ENST00000544796.1
ENST00000369644.1 |
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr1_-_161207986 | 0.17 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr10_-_103815874 | 0.17 |
ENST00000370033.4
ENST00000311122.5 |
C10orf76
|
chromosome 10 open reading frame 76 |
| chr11_-_71781096 | 0.17 |
ENST00000535087.1
ENST00000535838.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr1_-_151138323 | 0.16 |
ENST00000368908.5
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr22_+_25003568 | 0.16 |
ENST00000447416.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr2_+_44502630 | 0.16 |
ENST00000410056.3
ENST00000409741.1 ENST00000409229.3 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr11_-_118868682 | 0.16 |
ENST00000526453.1
|
RP11-110I1.12
|
RP11-110I1.12 |
| chr22_+_25003606 | 0.15 |
ENST00000432867.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr19_-_15235906 | 0.15 |
ENST00000600984.1
|
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr20_-_36156264 | 0.15 |
ENST00000445723.1
ENST00000414080.1 |
BLCAP
|
bladder cancer associated protein |
| chr18_+_54318566 | 0.15 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr3_-_125775629 | 0.15 |
ENST00000383598.2
|
SLC41A3
|
solute carrier family 41, member 3 |
| chr16_+_29840929 | 0.15 |
ENST00000566252.1
|
MVP
|
major vault protein |
| chr1_-_161208013 | 0.15 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr7_-_47621229 | 0.15 |
ENST00000434451.1
|
TNS3
|
tensin 3 |
| chr5_+_133842243 | 0.15 |
ENST00000515627.2
|
AC005355.2
|
AC005355.2 |
| chr8_-_128231299 | 0.14 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr14_+_23352374 | 0.14 |
ENST00000267396.4
ENST00000536884.1 |
REM2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr12_+_121416437 | 0.14 |
ENST00000402929.1
ENST00000535955.1 ENST00000538626.1 ENST00000543427.1 |
HNF1A
|
HNF1 homeobox A |
| chr19_+_10197463 | 0.14 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr14_-_51562037 | 0.14 |
ENST00000338969.5
|
TRIM9
|
tripartite motif containing 9 |
| chr10_-_21186144 | 0.14 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr17_-_7082668 | 0.14 |
ENST00000573083.1
ENST00000574388.1 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr11_+_119056178 | 0.14 |
ENST00000525131.1
ENST00000531114.1 ENST00000355547.5 ENST00000322712.4 |
PDZD3
|
PDZ domain containing 3 |
| chr2_+_241544834 | 0.14 |
ENST00000319838.5
ENST00000403859.1 ENST00000438013.2 |
GPR35
|
G protein-coupled receptor 35 |
| chr20_-_36156293 | 0.14 |
ENST00000373537.2
ENST00000414542.2 |
BLCAP
|
bladder cancer associated protein |
| chr16_-_88752889 | 0.13 |
ENST00000332281.5
|
SNAI3
|
snail family zinc finger 3 |
| chr19_+_4639514 | 0.13 |
ENST00000327473.4
|
TNFAIP8L1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr19_+_751122 | 0.13 |
ENST00000215582.6
|
MISP
|
mitotic spindle positioning |
| chr11_+_63137251 | 0.13 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chr12_+_56114189 | 0.13 |
ENST00000548082.1
|
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr11_-_63536113 | 0.12 |
ENST00000433688.1
ENST00000546282.2 |
C11orf95
RP11-466C23.4
|
chromosome 11 open reading frame 95 RP11-466C23.4 |
| chr1_+_43855560 | 0.12 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr22_-_31063782 | 0.12 |
ENST00000404885.1
ENST00000403268.1 ENST00000407308.1 ENST00000342474.4 ENST00000334679.3 |
DUSP18
|
dual specificity phosphatase 18 |
| chr19_-_10687983 | 0.12 |
ENST00000587069.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr2_-_61389168 | 0.11 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr2_+_219135115 | 0.11 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr6_+_131148538 | 0.11 |
ENST00000541421.2
|
SMLR1
|
small leucine-rich protein 1 |
| chr17_-_44896047 | 0.11 |
ENST00000225512.5
|
WNT3
|
wingless-type MMTV integration site family, member 3 |
| chr11_-_61658853 | 0.11 |
ENST00000525588.1
ENST00000540820.1 |
FADS3
|
fatty acid desaturase 3 |
| chr2_-_27603582 | 0.11 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr17_-_2614927 | 0.11 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr17_+_44039704 | 0.10 |
ENST00000420682.2
ENST00000415613.2 ENST00000571987.1 ENST00000574436.1 ENST00000431008.3 |
MAPT
|
microtubule-associated protein tau |
| chr12_+_47617284 | 0.10 |
ENST00000549630.1
ENST00000551777.1 |
PCED1B
|
PC-esterase domain containing 1B |
| chr16_+_21244986 | 0.10 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr17_-_2615031 | 0.10 |
ENST00000576885.1
ENST00000574426.2 |
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr1_-_43751230 | 0.10 |
ENST00000523677.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr14_-_51561784 | 0.10 |
ENST00000360392.4
|
TRIM9
|
tripartite motif containing 9 |
| chr19_-_48867291 | 0.10 |
ENST00000435956.3
|
TMEM143
|
transmembrane protein 143 |
| chr6_+_31949801 | 0.09 |
ENST00000428956.2
ENST00000498271.1 |
C4A
|
complement component 4A (Rodgers blood group) |
| chr9_-_27005686 | 0.09 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr5_-_42812143 | 0.09 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr14_-_76447336 | 0.09 |
ENST00000556285.1
|
TGFB3
|
transforming growth factor, beta 3 |
| chr7_+_45927956 | 0.09 |
ENST00000275525.3
ENST00000457280.1 |
IGFBP1
|
insulin-like growth factor binding protein 1 |
| chr19_-_42758040 | 0.09 |
ENST00000593944.1
|
ERF
|
Ets2 repressor factor |
| chr2_+_48796120 | 0.09 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr2_+_97454321 | 0.09 |
ENST00000540067.1
|
CNNM4
|
cyclin M4 |
| chr1_+_199996733 | 0.09 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr12_-_51422017 | 0.09 |
ENST00000394904.3
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr5_+_79950463 | 0.09 |
ENST00000265081.6
|
MSH3
|
mutS homolog 3 |
| chr22_+_25003626 | 0.09 |
ENST00000451366.1
ENST00000406383.2 ENST00000428855.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr13_+_50656307 | 0.09 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr14_+_100485712 | 0.08 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr19_+_7660716 | 0.08 |
ENST00000160298.4
ENST00000446248.2 |
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr19_-_41256207 | 0.08 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr14_+_77648167 | 0.08 |
ENST00000554346.1
ENST00000298351.4 |
TMEM63C
|
transmembrane protein 63C |
| chr6_+_31926857 | 0.08 |
ENST00000375394.2
ENST00000544581.1 |
SKIV2L
|
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr18_+_42260059 | 0.08 |
ENST00000426838.4
|
SETBP1
|
SET binding protein 1 |
| chr7_-_15601595 | 0.08 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr19_+_11546153 | 0.08 |
ENST00000591946.1
ENST00000252455.2 ENST00000412601.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr2_-_61389240 | 0.08 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr12_-_118810688 | 0.08 |
ENST00000542532.1
ENST00000392533.3 |
TAOK3
|
TAO kinase 3 |
| chr1_-_11863171 | 0.08 |
ENST00000376592.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr17_+_61554413 | 0.08 |
ENST00000538928.1
ENST00000290866.4 ENST00000428043.1 |
ACE
|
angiotensin I converting enzyme |
| chr16_-_87970122 | 0.07 |
ENST00000309893.2
|
CA5A
|
carbonic anhydrase VA, mitochondrial |
| chr14_+_32476072 | 0.07 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chr17_-_17494972 | 0.07 |
ENST00000435340.2
ENST00000255389.5 ENST00000395781.2 |
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr2_-_62733476 | 0.07 |
ENST00000335390.5
|
TMEM17
|
transmembrane protein 17 |
| chr19_+_11546440 | 0.07 |
ENST00000589126.1
ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr1_-_177939348 | 0.07 |
ENST00000464631.2
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr5_+_14664762 | 0.07 |
ENST00000284274.4
|
FAM105B
|
family with sequence similarity 105, member B |
| chr5_-_1801408 | 0.06 |
ENST00000505818.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr10_+_104404218 | 0.06 |
ENST00000302424.7
|
TRIM8
|
tripartite motif containing 8 |
| chr10_+_81065975 | 0.06 |
ENST00000446377.2
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chr6_+_31982539 | 0.06 |
ENST00000435363.2
ENST00000425700.2 |
C4B
|
complement component 4B (Chido blood group) |
| chr22_+_27068766 | 0.06 |
ENST00000435162.1
ENST00000437071.1 ENST00000440816.1 ENST00000421253.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr19_+_39881951 | 0.06 |
ENST00000315588.5
ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29
|
mediator complex subunit 29 |
| chr3_+_184033551 | 0.06 |
ENST00000456033.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr6_-_32145861 | 0.06 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr20_+_62694461 | 0.06 |
ENST00000343484.5
ENST00000395053.3 |
TCEA2
|
transcription elongation factor A (SII), 2 |
| chr20_-_44485835 | 0.06 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr15_+_63340858 | 0.05 |
ENST00000560615.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr1_-_41131326 | 0.05 |
ENST00000372684.3
|
RIMS3
|
regulating synaptic membrane exocytosis 3 |
| chr2_-_222436988 | 0.05 |
ENST00000409854.1
ENST00000281821.2 ENST00000392071.4 ENST00000443796.1 |
EPHA4
|
EPH receptor A4 |
| chr2_-_219134822 | 0.05 |
ENST00000444053.1
ENST00000248450.4 |
AAMP
|
angio-associated, migratory cell protein |
| chr4_-_109541539 | 0.05 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr16_+_24857552 | 0.05 |
ENST00000568579.1
ENST00000567758.1 ENST00000569071.1 ENST00000539472.1 |
SLC5A11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr1_-_60392452 | 0.05 |
ENST00000371204.3
|
CYP2J2
|
cytochrome P450, family 2, subfamily J, polypeptide 2 |
| chr6_-_33385854 | 0.05 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr9_-_136605042 | 0.05 |
ENST00000371872.4
ENST00000298628.5 ENST00000422262.2 |
SARDH
|
sarcosine dehydrogenase |
| chr10_-_116444371 | 0.05 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr1_+_199996702 | 0.04 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr8_-_98290087 | 0.04 |
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5 |
| chr5_-_173043591 | 0.04 |
ENST00000285908.5
ENST00000480951.1 ENST00000311086.4 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr18_+_54318616 | 0.04 |
ENST00000254442.3
|
WDR7
|
WD repeat domain 7 |
| chr14_-_23446021 | 0.04 |
ENST00000553592.1
|
AJUBA
|
ajuba LIM protein |
| chr15_-_41408409 | 0.04 |
ENST00000361937.3
|
INO80
|
INO80 complex subunit |
| chr17_+_7591639 | 0.04 |
ENST00000396463.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr12_-_54121212 | 0.04 |
ENST00000548263.1
ENST00000430117.2 ENST00000550804.1 ENST00000549173.1 ENST00000551900.1 ENST00000546619.1 ENST00000548177.1 ENST00000549349.1 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr12_-_7125770 | 0.04 |
ENST00000261407.4
|
LPCAT3
|
lysophosphatidylcholine acyltransferase 3 |
| chr2_-_159313214 | 0.04 |
ENST00000409889.1
ENST00000283233.5 ENST00000536771.1 |
CCDC148
|
coiled-coil domain containing 148 |
| chr1_-_15911510 | 0.04 |
ENST00000375826.3
|
AGMAT
|
agmatine ureohydrolase (agmatinase) |
| chr6_+_147527103 | 0.03 |
ENST00000179882.6
|
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr10_+_96522361 | 0.03 |
ENST00000371321.3
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19 |
| chr5_+_1801503 | 0.03 |
ENST00000274137.5
ENST00000469176.1 |
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) |
| chr19_+_39616410 | 0.03 |
ENST00000602004.1
ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr3_-_141868293 | 0.03 |
ENST00000317104.7
ENST00000494358.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr22_+_30163340 | 0.03 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr19_+_1205740 | 0.03 |
ENST00000326873.7
|
STK11
|
serine/threonine kinase 11 |
| chr5_-_102455801 | 0.03 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr5_+_176811431 | 0.03 |
ENST00000512593.1
ENST00000324417.5 |
SLC34A1
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr1_-_161207953 | 0.03 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr19_+_11546093 | 0.03 |
ENST00000591462.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr2_+_73441350 | 0.03 |
ENST00000389501.4
|
SMYD5
|
SMYD family member 5 |
| chr10_+_96443378 | 0.03 |
ENST00000285979.6
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr11_-_71750865 | 0.02 |
ENST00000544129.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr3_+_108541545 | 0.02 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr3_+_108541608 | 0.02 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr1_-_151138422 | 0.02 |
ENST00000440902.2
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr3_-_49851313 | 0.02 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr3_+_191046810 | 0.02 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr14_+_64680854 | 0.02 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr14_+_101295638 | 0.02 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr4_+_144312659 | 0.02 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr19_-_39881777 | 0.02 |
ENST00000595564.1
ENST00000221265.3 |
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr7_+_143080063 | 0.01 |
ENST00000446634.1
|
ZYX
|
zyxin |
| chr15_+_50716576 | 0.01 |
ENST00000560297.1
ENST00000307179.4 ENST00000396444.3 ENST00000433963.1 ENST00000425032.3 |
USP8
|
ubiquitin specific peptidase 8 |
| chr17_+_1646130 | 0.01 |
ENST00000453066.1
ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr6_+_31950150 | 0.01 |
ENST00000537134.1
|
C4A
|
complement component 4A (Rodgers blood group) |
| chr3_-_141868357 | 0.01 |
ENST00000489671.1
ENST00000475734.1 ENST00000467072.1 ENST00000499676.2 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chrX_-_77041685 | 0.01 |
ENST00000373344.5
ENST00000395603.3 |
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked |
| chr15_+_63340734 | 0.01 |
ENST00000560959.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr3_-_52002194 | 0.01 |
ENST00000466412.1
|
PCBP4
|
poly(rC) binding protein 4 |
| chr14_-_39572345 | 0.01 |
ENST00000548032.2
ENST00000556092.1 ENST00000557280.1 ENST00000545328.2 ENST00000553970.1 |
SEC23A
|
Sec23 homolog A (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HNF4A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.2 | 0.5 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.1 | 0.5 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.2 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.2 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.1 | 0.2 | GO:0060748 | negative regulation of integrin biosynthetic process(GO:0045720) tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.1 | 0.2 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 0.4 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.2 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.1 | 0.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.4 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.0 | 0.5 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.2 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.6 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.0 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.2 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.1 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.6 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.6 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.0 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.1 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.0 | 0.9 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.2 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.3 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.6 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 0.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.4 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.4 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0000406 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0030249 | cyclase regulator activity(GO:0010851) guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.0 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |