Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
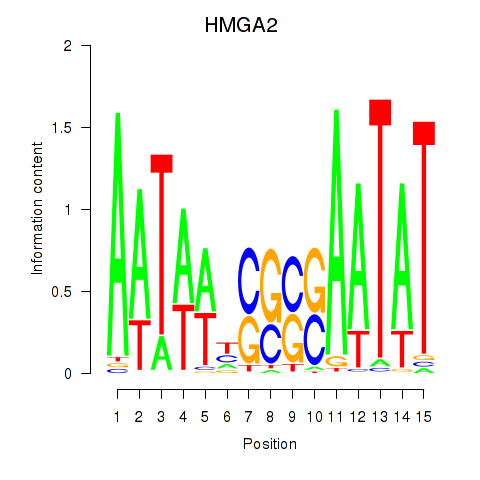
Results for HMGA2
Z-value: 0.28
Transcription factors associated with HMGA2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMGA2
|
ENSG00000149948.9 | high mobility group AT-hook 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMGA2 | hg19_v2_chr12_+_66218212_66218244 | -0.95 | 5.1e-02 | Click! |
Activity profile of HMGA2 motif
Sorted Z-values of HMGA2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_65996599 | 0.11 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr2_-_151395525 | 0.09 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr10_-_36813162 | 0.08 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr15_+_30375158 | 0.08 |
ENST00000341650.6
ENST00000567927.1 |
GOLGA8J
|
golgin A8 family, member J |
| chr9_-_115480303 | 0.07 |
ENST00000374234.1
ENST00000374238.1 ENST00000374236.1 ENST00000374242.4 |
INIP
|
INTS3 and NABP interacting protein |
| chrX_-_128782722 | 0.07 |
ENST00000427399.1
|
APLN
|
apelin |
| chr12_-_8765446 | 0.07 |
ENST00000537228.1
ENST00000229335.6 |
AICDA
|
activation-induced cytidine deaminase |
| chr1_+_227751231 | 0.06 |
ENST00000343776.5
ENST00000608949.1 ENST00000397097.3 |
ZNF678
|
zinc finger protein 678 |
| chr1_-_43638168 | 0.06 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr2_-_114461655 | 0.06 |
ENST00000424612.1
|
AC017074.2
|
AC017074.2 |
| chr4_+_40058411 | 0.06 |
ENST00000261435.6
ENST00000515550.1 |
N4BP2
|
NEDD4 binding protein 2 |
| chr10_-_101673782 | 0.06 |
ENST00000422692.1
|
DNMBP
|
dynamin binding protein |
| chr15_+_43885252 | 0.05 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr19_+_44455368 | 0.05 |
ENST00000591168.1
ENST00000587682.1 ENST00000251269.5 |
ZNF221
|
zinc finger protein 221 |
| chr10_+_35456444 | 0.05 |
ENST00000361599.4
|
CREM
|
cAMP responsive element modulator |
| chr17_+_73230799 | 0.05 |
ENST00000579838.1
|
NUP85
|
nucleoporin 85kDa |
| chr7_-_102985035 | 0.05 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr14_-_94759595 | 0.04 |
ENST00000261994.4
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr12_-_21757774 | 0.04 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver) |
| chr12_+_69979113 | 0.04 |
ENST00000299300.6
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chrX_+_2670066 | 0.04 |
ENST00000381174.5
ENST00000419513.2 ENST00000426774.1 |
XG
|
Xg blood group |
| chr1_-_67600639 | 0.04 |
ENST00000544837.1
ENST00000603691.1 |
C1orf141
|
chromosome 1 open reading frame 141 |
| chr5_-_102455801 | 0.04 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr9_+_108463234 | 0.04 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chrX_-_7895479 | 0.04 |
ENST00000381042.4
|
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr1_-_226374373 | 0.04 |
ENST00000366812.5
|
ACBD3
|
acyl-CoA binding domain containing 3 |
| chr15_+_43985084 | 0.04 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr14_-_94759408 | 0.04 |
ENST00000554723.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr12_-_122750957 | 0.04 |
ENST00000451053.2
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae) |
| chr14_-_94759361 | 0.04 |
ENST00000393096.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr9_+_104296122 | 0.04 |
ENST00000389120.3
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr11_+_65122216 | 0.04 |
ENST00000309880.5
|
TIGD3
|
tigger transposable element derived 3 |
| chr20_+_54967663 | 0.04 |
ENST00000452950.1
|
CSTF1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa |
| chr14_+_45553296 | 0.04 |
ENST00000355765.6
ENST00000553605.1 |
PRPF39
|
pre-mRNA processing factor 39 |
| chr15_+_84904525 | 0.04 |
ENST00000510439.2
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chr6_-_127664683 | 0.04 |
ENST00000528402.1
ENST00000454591.2 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr1_+_28836561 | 0.03 |
ENST00000419074.1
|
RCC1
|
regulator of chromosome condensation 1 |
| chr17_-_48785216 | 0.03 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr15_-_83018198 | 0.03 |
ENST00000557886.1
|
RP13-996F3.4
|
golgin A6 family-like 19 |
| chr3_-_123680246 | 0.03 |
ENST00000488653.2
|
CCDC14
|
coiled-coil domain containing 14 |
| chr15_-_82939157 | 0.03 |
ENST00000559949.1
|
RP13-996F3.5
|
golgin A6 family-like 18 |
| chr1_-_223988426 | 0.03 |
ENST00000391879.2
|
TP53BP2
|
tumor protein p53 binding protein, 2 |
| chr4_+_25378912 | 0.03 |
ENST00000510092.1
ENST00000505991.1 |
ANAPC4
|
anaphase promoting complex subunit 4 |
| chr6_+_116892641 | 0.03 |
ENST00000487832.2
ENST00000518117.1 |
RWDD1
|
RWD domain containing 1 |
| chrX_+_117480036 | 0.03 |
ENST00000371822.5
ENST00000254029.3 ENST00000371825.3 |
WDR44
|
WD repeat domain 44 |
| chr21_+_38792602 | 0.03 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr15_+_82722225 | 0.03 |
ENST00000300515.8
|
GOLGA6L9
|
golgin A6 family-like 9 |
| chr20_+_46130671 | 0.03 |
ENST00000371998.3
ENST00000371997.3 |
NCOA3
|
nuclear receptor coactivator 3 |
| chr15_+_83098710 | 0.03 |
ENST00000561062.1
ENST00000358583.3 |
GOLGA6L9
|
golgin A6 family-like 20 |
| chr15_+_28623784 | 0.03 |
ENST00000526619.2
ENST00000337838.7 ENST00000532622.2 |
GOLGA8F
|
golgin A8 family, member F |
| chr3_+_193853927 | 0.03 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr12_+_69979210 | 0.03 |
ENST00000544368.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr19_-_11456872 | 0.02 |
ENST00000586218.1
|
TMEM205
|
transmembrane protein 205 |
| chr12_-_10573149 | 0.02 |
ENST00000381904.2
ENST00000381903.2 ENST00000396439.2 |
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr7_-_102985288 | 0.02 |
ENST00000379263.3
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr8_-_105479270 | 0.02 |
ENST00000521573.2
ENST00000351513.2 |
DPYS
|
dihydropyrimidinase |
| chrX_-_23926004 | 0.02 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr20_+_46130619 | 0.02 |
ENST00000372004.3
|
NCOA3
|
nuclear receptor coactivator 3 |
| chr6_-_136571400 | 0.02 |
ENST00000418509.2
ENST00000420702.1 ENST00000451457.2 |
MTFR2
|
mitochondrial fission regulator 2 |
| chr1_+_197886461 | 0.02 |
ENST00000367388.3
ENST00000337020.2 ENST00000367387.4 |
LHX9
|
LIM homeobox 9 |
| chr15_-_32747835 | 0.02 |
ENST00000509311.2
ENST00000414865.2 |
GOLGA8O
|
golgin A8 family, member O |
| chr2_+_172543967 | 0.02 |
ENST00000534253.2
ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr2_+_13677795 | 0.02 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr1_+_179263308 | 0.02 |
ENST00000426956.1
|
SOAT1
|
sterol O-acyltransferase 1 |
| chr14_+_99947715 | 0.02 |
ENST00000389879.5
ENST00000557441.1 ENST00000555049.1 ENST00000555842.1 |
CCNK
|
cyclin K |
| chrX_-_135962876 | 0.02 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr6_+_88106840 | 0.02 |
ENST00000369570.4
|
C6orf164
|
chromosome 6 open reading frame 164 |
| chr14_+_88852059 | 0.02 |
ENST00000045347.7
|
SPATA7
|
spermatogenesis associated 7 |
| chr13_+_51483814 | 0.01 |
ENST00000336617.3
ENST00000422660.1 |
RNASEH2B
|
ribonuclease H2, subunit B |
| chr2_+_170683979 | 0.01 |
ENST00000418381.1
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr12_-_122751002 | 0.01 |
ENST00000267199.4
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae) |
| chr10_+_86184676 | 0.01 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr10_+_96305535 | 0.01 |
ENST00000419900.1
ENST00000348459.5 ENST00000394045.1 ENST00000394044.1 ENST00000394036.1 |
HELLS
|
helicase, lymphoid-specific |
| chr11_+_67250490 | 0.01 |
ENST00000528641.2
ENST00000279146.3 |
AIP
|
aryl hydrocarbon receptor interacting protein |
| chr1_+_104104379 | 0.01 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr18_-_14132422 | 0.01 |
ENST00000589498.1
ENST00000590202.1 |
ZNF519
|
zinc finger protein 519 |
| chr2_-_130031335 | 0.01 |
ENST00000375987.3
|
AC079586.1
|
AC079586.1 |
| chr15_+_30427352 | 0.01 |
ENST00000569052.1
|
GOLGA8T
|
golgin A8 family, member T |
| chr17_-_26220366 | 0.01 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr16_+_53483983 | 0.01 |
ENST00000544545.1
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr20_+_46130601 | 0.01 |
ENST00000341724.6
|
NCOA3
|
nuclear receptor coactivator 3 |
| chr2_+_172543919 | 0.01 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr2_+_172544011 | 0.01 |
ENST00000508530.1
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr13_+_50656307 | 0.01 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr9_-_86955598 | 0.01 |
ENST00000376238.4
|
SLC28A3
|
solute carrier family 28 (concentrative nucleoside transporter), member 3 |
| chr12_-_10541575 | 0.01 |
ENST00000540818.1
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr9_-_34397800 | 0.01 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr2_+_177134134 | 0.01 |
ENST00000249442.6
ENST00000392529.2 ENST00000443241.1 |
MTX2
|
metaxin 2 |
| chr15_+_58702742 | 0.00 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chrX_+_1710484 | 0.00 |
ENST00000313871.3
ENST00000381261.3 |
AKAP17A
|
A kinase (PRKA) anchor protein 17A |
| chr14_+_37641012 | 0.00 |
ENST00000556667.1
|
SLC25A21-AS1
|
SLC25A21 antisense RNA 1 |
| chr11_+_69455855 | 0.00 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr6_-_127664736 | 0.00 |
ENST00000368291.2
ENST00000309620.9 ENST00000454859.3 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr3_+_137728842 | 0.00 |
ENST00000183605.5
|
CLDN18
|
claudin 18 |
| chr19_-_38146289 | 0.00 |
ENST00000392144.1
ENST00000591444.1 ENST00000351218.2 ENST00000587809.1 |
ZFP30
|
ZFP30 zinc finger protein |
| chr3_-_157221128 | 0.00 |
ENST00000392833.2
ENST00000362010.2 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr18_+_11851383 | 0.00 |
ENST00000526991.2
|
CHMP1B
|
charged multivesicular body protein 1B |
| chr6_-_32908765 | 0.00 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chrX_-_77150985 | 0.00 |
ENST00000358075.6
|
MAGT1
|
magnesium transporter 1 |
| chrX_-_135962923 | 0.00 |
ENST00000565438.1
|
RBMX
|
RNA binding motif protein, X-linked |
| chr1_+_227751281 | 0.00 |
ENST00000440339.1
|
ZNF678
|
zinc finger protein 678 |
| chr16_+_66442411 | 0.00 |
ENST00000499966.1
|
LINC00920
|
long intergenic non-protein coding RNA 920 |
| chr15_-_70994612 | 0.00 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr18_-_21017817 | 0.00 |
ENST00000542162.1
ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241
|
transmembrane protein 241 |
| chr10_-_62332357 | 0.00 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMGA2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.1 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |