Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
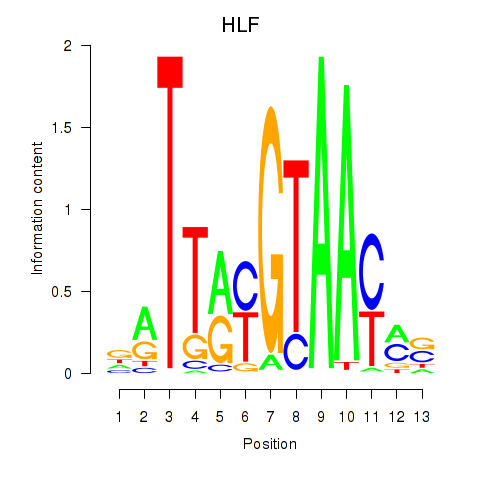
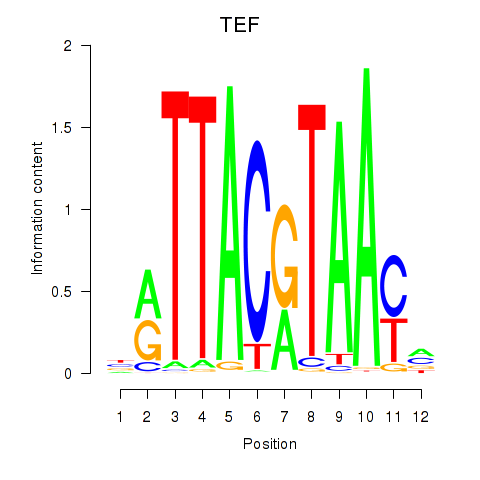
Results for HLF_TEF
Z-value: 0.97
Transcription factors associated with HLF_TEF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HLF
|
ENSG00000108924.9 | HLF transcription factor, PAR bZIP family member |
|
TEF
|
ENSG00000167074.10 | TEF transcription factor, PAR bZIP family member |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HLF | hg19_v2_chr17_+_53345080_53345177 | -1.00 | 1.5e-03 | Click! |
| TEF | hg19_v2_chr22_+_41763274_41763337 | 0.08 | 9.2e-01 | Click! |
Activity profile of HLF_TEF motif
Sorted Z-values of HLF_TEF motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_34665983 | 1.37 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr20_+_4667094 | 0.67 |
ENST00000424424.1
ENST00000457586.1 |
PRNP
|
prion protein |
| chr12_-_46121554 | 0.59 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr14_+_78227105 | 0.57 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr8_+_94752349 | 0.42 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr1_+_7844312 | 0.41 |
ENST00000377541.1
|
PER3
|
period circadian clock 3 |
| chr13_-_29292956 | 0.41 |
ENST00000266943.6
|
SLC46A3
|
solute carrier family 46, member 3 |
| chr8_-_27469383 | 0.41 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr8_-_102218292 | 0.40 |
ENST00000518336.1
ENST00000520454.1 |
ZNF706
|
zinc finger protein 706 |
| chr12_-_12714006 | 0.39 |
ENST00000541207.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr12_-_12714025 | 0.38 |
ENST00000539940.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr8_+_19536083 | 0.37 |
ENST00000519803.1
|
RP11-1105O14.1
|
RP11-1105O14.1 |
| chr7_-_83824449 | 0.36 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_-_41322332 | 0.36 |
ENST00000590740.1
|
RP11-242D8.1
|
RP11-242D8.1 |
| chr12_+_52431016 | 0.31 |
ENST00000553200.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr7_+_90032821 | 0.31 |
ENST00000427904.1
|
CLDN12
|
claudin 12 |
| chr2_+_65663812 | 0.30 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr17_+_76037081 | 0.29 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr16_-_54962704 | 0.23 |
ENST00000502066.2
ENST00000560912.1 ENST00000558952.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr17_+_70036164 | 0.23 |
ENST00000602013.1
|
AC007461.1
|
Uncharacterized protein |
| chr1_-_161207986 | 0.22 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr14_+_102276192 | 0.22 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr17_-_73178599 | 0.21 |
ENST00000578238.1
|
SUMO2
|
small ubiquitin-like modifier 2 |
| chr14_+_96722152 | 0.20 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr3_+_182983126 | 0.20 |
ENST00000481531.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr4_+_26344754 | 0.19 |
ENST00000515573.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr14_-_23284703 | 0.19 |
ENST00000555911.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr14_+_96722539 | 0.19 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr14_-_23285069 | 0.19 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr3_+_141103634 | 0.19 |
ENST00000507722.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr3_-_196910477 | 0.18 |
ENST00000447466.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr19_+_1450112 | 0.18 |
ENST00000590469.1
ENST00000233607.2 ENST00000238483.4 ENST00000590877.1 |
APC2
|
adenomatosis polyposis coli 2 |
| chr14_-_23284675 | 0.18 |
ENST00000555959.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr16_+_70680439 | 0.18 |
ENST00000288098.2
|
IL34
|
interleukin 34 |
| chr5_-_140013241 | 0.18 |
ENST00000519715.1
|
CD14
|
CD14 molecule |
| chr11_-_33913708 | 0.18 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr14_+_32476072 | 0.18 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chr7_+_20686946 | 0.17 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr4_+_96012585 | 0.17 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr10_+_86004802 | 0.17 |
ENST00000359452.4
ENST00000358110.5 ENST00000372092.3 |
RGR
|
retinal G protein coupled receptor |
| chr17_-_685493 | 0.16 |
ENST00000536578.1
ENST00000301328.5 ENST00000576419.1 |
GLOD4
|
glyoxalase domain containing 4 |
| chr1_-_226496772 | 0.16 |
ENST00000359525.2
ENST00000460719.1 |
LIN9
|
lin-9 homolog (C. elegans) |
| chr7_-_111428957 | 0.15 |
ENST00000417165.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr8_+_42396936 | 0.15 |
ENST00000416469.2
|
SMIM19
|
small integral membrane protein 19 |
| chr1_+_40839369 | 0.15 |
ENST00000372718.3
|
SMAP2
|
small ArfGAP2 |
| chr12_-_88423164 | 0.14 |
ENST00000298699.2
ENST00000550553.1 |
C12orf50
|
chromosome 12 open reading frame 50 |
| chr12_+_48592134 | 0.14 |
ENST00000595310.1
|
DKFZP779L1853
|
DKFZP779L1853 |
| chr3_-_45883558 | 0.14 |
ENST00000445698.1
ENST00000296135.6 |
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr7_+_23146271 | 0.14 |
ENST00000545771.1
|
KLHL7
|
kelch-like family member 7 |
| chr19_+_42254885 | 0.14 |
ENST00000595740.1
|
CEACAM6
|
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) |
| chr11_+_58938903 | 0.13 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr2_-_240230890 | 0.13 |
ENST00000446876.1
|
HDAC4
|
histone deacetylase 4 |
| chr19_-_1155118 | 0.13 |
ENST00000590998.1
|
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr2_-_113594279 | 0.12 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr8_+_110552337 | 0.12 |
ENST00000337573.5
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr2_-_167232484 | 0.12 |
ENST00000375387.4
ENST00000303354.6 ENST00000409672.1 |
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit |
| chr1_+_154378049 | 0.12 |
ENST00000512471.1
|
IL6R
|
interleukin 6 receptor |
| chr10_-_94050820 | 0.12 |
ENST00000265997.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr17_-_685559 | 0.12 |
ENST00000301329.6
|
GLOD4
|
glyoxalase domain containing 4 |
| chr9_+_110045418 | 0.11 |
ENST00000419616.1
|
RAD23B
|
RAD23 homolog B (S. cerevisiae) |
| chr1_+_144339738 | 0.11 |
ENST00000538264.1
|
AL592284.1
|
Protein LOC642441 |
| chr1_-_161207875 | 0.11 |
ENST00000512372.1
ENST00000437437.2 ENST00000442691.2 ENST00000412844.2 ENST00000428574.2 ENST00000505005.1 ENST00000508740.1 ENST00000508387.1 ENST00000504010.1 ENST00000511676.1 ENST00000502985.1 ENST00000367981.3 ENST00000515621.1 ENST00000511944.1 ENST00000511748.1 ENST00000367984.4 ENST00000367985.3 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr8_-_102217515 | 0.11 |
ENST00000520347.1
ENST00000523922.1 ENST00000520984.1 |
ZNF706
|
zinc finger protein 706 |
| chr5_-_171404730 | 0.11 |
ENST00000518752.1
|
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr16_-_30102547 | 0.11 |
ENST00000279386.2
|
TBX6
|
T-box 6 |
| chr17_-_61850894 | 0.11 |
ENST00000403162.3
ENST00000582252.1 ENST00000225726.5 |
CCDC47
|
coiled-coil domain containing 47 |
| chr13_-_46626847 | 0.11 |
ENST00000242848.4
ENST00000282007.3 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr3_+_169684553 | 0.10 |
ENST00000337002.4
ENST00000480708.1 |
SEC62
|
SEC62 homolog (S. cerevisiae) |
| chr7_-_141401951 | 0.10 |
ENST00000536163.1
|
KIAA1147
|
KIAA1147 |
| chr14_+_67708344 | 0.10 |
ENST00000557237.1
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr3_+_160559931 | 0.10 |
ENST00000464260.1
ENST00000295839.9 |
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr14_-_67826538 | 0.10 |
ENST00000553687.1
|
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr18_+_32621324 | 0.10 |
ENST00000300249.5
ENST00000538170.2 ENST00000588910.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr14_-_23285011 | 0.10 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr7_-_44887620 | 0.10 |
ENST00000349299.3
ENST00000521529.1 ENST00000308153.4 ENST00000350771.3 ENST00000222690.6 ENST00000381124.5 ENST00000437072.1 ENST00000446531.1 |
H2AFV
|
H2A histone family, member V |
| chr19_+_52076425 | 0.10 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chr1_-_161208013 | 0.09 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr5_-_179050066 | 0.09 |
ENST00000329433.6
ENST00000510411.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr1_+_174933899 | 0.09 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr3_+_63953415 | 0.09 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr21_+_45138941 | 0.09 |
ENST00000398081.1
ENST00000468090.1 ENST00000291565.4 |
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr1_+_196621156 | 0.09 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr6_-_86353510 | 0.09 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr19_-_4065730 | 0.08 |
ENST00000601588.1
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr1_-_161087802 | 0.08 |
ENST00000368010.3
|
PFDN2
|
prefoldin subunit 2 |
| chr4_+_96012614 | 0.08 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr12_-_101801505 | 0.08 |
ENST00000539055.1
ENST00000551688.1 ENST00000551671.1 ENST00000261636.8 |
ARL1
|
ADP-ribosylation factor-like 1 |
| chr11_-_33795893 | 0.08 |
ENST00000526785.1
ENST00000534136.1 ENST00000265651.3 ENST00000530401.1 ENST00000448981.2 |
FBXO3
|
F-box protein 3 |
| chr18_-_55288973 | 0.08 |
ENST00000423481.2
ENST00000587194.1 ENST00000591599.1 ENST00000588661.1 |
NARS
|
asparaginyl-tRNA synthetase |
| chr4_-_74088800 | 0.08 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr3_-_196910721 | 0.08 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr16_+_3507985 | 0.07 |
ENST00000421765.3
ENST00000360862.5 ENST00000414063.2 ENST00000610180.1 ENST00000608993.1 |
NAA60
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit N-alpha-acetyltransferase 60 |
| chr7_-_73153178 | 0.07 |
ENST00000437775.2
ENST00000222800.3 |
ABHD11
|
abhydrolase domain containing 11 |
| chr1_+_25757376 | 0.07 |
ENST00000399766.3
ENST00000399763.3 ENST00000374343.4 |
TMEM57
|
transmembrane protein 57 |
| chr6_-_30043539 | 0.07 |
ENST00000376751.3
ENST00000244360.6 |
RNF39
|
ring finger protein 39 |
| chr13_-_28545276 | 0.07 |
ENST00000381020.7
|
CDX2
|
caudal type homeobox 2 |
| chr1_+_75594119 | 0.07 |
ENST00000294638.5
|
LHX8
|
LIM homeobox 8 |
| chr4_-_185395191 | 0.07 |
ENST00000510814.1
ENST00000507523.1 ENST00000506230.1 |
IRF2
|
interferon regulatory factor 2 |
| chr5_+_172332220 | 0.07 |
ENST00000518247.1
ENST00000326654.2 |
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr10_-_14372870 | 0.07 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr12_+_52430894 | 0.07 |
ENST00000546842.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr14_+_60716159 | 0.06 |
ENST00000325658.3
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr1_+_90308981 | 0.06 |
ENST00000527156.1
|
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr11_-_105948040 | 0.06 |
ENST00000534815.1
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr9_+_71820057 | 0.06 |
ENST00000539225.1
|
TJP2
|
tight junction protein 2 |
| chr8_-_103424916 | 0.06 |
ENST00000220959.4
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr2_-_100106419 | 0.06 |
ENST00000393445.3
ENST00000258428.3 |
REV1
|
REV1, polymerase (DNA directed) |
| chr1_+_92632542 | 0.05 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr2_+_102608306 | 0.05 |
ENST00000332549.3
|
IL1R2
|
interleukin 1 receptor, type II |
| chr8_-_103424986 | 0.05 |
ENST00000521922.1
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr16_+_70332956 | 0.05 |
ENST00000288071.6
ENST00000393657.2 ENST00000355992.3 ENST00000567706.1 |
DDX19B
RP11-529K1.3
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19B Uncharacterized protein |
| chr3_-_48130707 | 0.05 |
ENST00000360240.6
ENST00000383737.4 |
MAP4
|
microtubule-associated protein 4 |
| chr2_+_192543694 | 0.05 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr14_+_102276132 | 0.05 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr5_-_58882219 | 0.05 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr5_-_54830871 | 0.05 |
ENST00000307259.8
|
PPAP2A
|
phosphatidic acid phosphatase type 2A |
| chr8_-_27468717 | 0.05 |
ENST00000520796.1
ENST00000520491.1 |
CLU
|
clusterin |
| chr3_+_188817891 | 0.05 |
ENST00000412373.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr5_+_95998070 | 0.05 |
ENST00000421689.2
ENST00000510756.1 ENST00000512620.1 |
CAST
|
calpastatin |
| chr8_-_102216925 | 0.05 |
ENST00000517844.1
|
ZNF706
|
zinc finger protein 706 |
| chr20_+_16729003 | 0.05 |
ENST00000246081.2
|
OTOR
|
otoraplin |
| chr1_-_101491319 | 0.05 |
ENST00000342173.7
ENST00000488176.1 ENST00000370109.3 |
DPH5
|
diphthamide biosynthesis 5 |
| chr10_-_65028817 | 0.04 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr16_-_54962415 | 0.04 |
ENST00000501177.3
ENST00000559598.2 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr8_-_112248400 | 0.04 |
ENST00000519506.1
ENST00000522778.1 |
RP11-946L20.4
|
RP11-946L20.4 |
| chr4_-_21950356 | 0.04 |
ENST00000447367.2
ENST00000382152.2 |
KCNIP4
|
Kv channel interacting protein 4 |
| chr11_+_35201826 | 0.04 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr8_+_90914073 | 0.04 |
ENST00000297438.2
|
OSGIN2
|
oxidative stress induced growth inhibitor family member 2 |
| chr3_+_157154578 | 0.04 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr1_-_156721389 | 0.04 |
ENST00000537739.1
|
HDGF
|
hepatoma-derived growth factor |
| chr10_-_65028938 | 0.04 |
ENST00000402544.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr1_-_161207953 | 0.04 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr3_-_48130314 | 0.03 |
ENST00000439356.1
ENST00000395734.3 ENST00000426837.2 |
MAP4
|
microtubule-associated protein 4 |
| chr17_+_61851504 | 0.03 |
ENST00000359353.5
ENST00000389924.2 |
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr1_+_154377669 | 0.03 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr7_-_83824169 | 0.03 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr19_-_41388657 | 0.03 |
ENST00000301146.4
ENST00000291764.3 |
CYP2A7
|
cytochrome P450, family 2, subfamily A, polypeptide 7 |
| chr8_-_103425047 | 0.03 |
ENST00000520539.1
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr7_+_90032667 | 0.03 |
ENST00000496677.1
ENST00000287916.4 ENST00000535571.1 ENST00000394604.1 ENST00000394605.2 |
CLDN12
|
claudin 12 |
| chr3_+_148447887 | 0.03 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr17_+_61851157 | 0.03 |
ENST00000578681.1
ENST00000583590.1 |
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr5_-_138534071 | 0.03 |
ENST00000394817.2
|
SIL1
|
SIL1 nucleotide exchange factor |
| chr19_-_35992780 | 0.03 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr2_+_46769798 | 0.03 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr22_-_29711704 | 0.03 |
ENST00000216101.6
|
RASL10A
|
RAS-like, family 10, member A |
| chr5_+_95997885 | 0.03 |
ENST00000511097.2
|
CAST
|
calpastatin |
| chr1_-_167883327 | 0.03 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr7_-_15601595 | 0.03 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr2_+_183989083 | 0.03 |
ENST00000295119.4
|
NUP35
|
nucleoporin 35kDa |
| chr21_-_30365136 | 0.03 |
ENST00000361371.5
ENST00000389194.2 ENST00000389195.2 |
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr1_+_45212051 | 0.03 |
ENST00000372222.3
|
KIF2C
|
kinesin family member 2C |
| chr11_-_62313090 | 0.02 |
ENST00000528508.1
ENST00000533365.1 |
AHNAK
|
AHNAK nucleoprotein |
| chr6_+_97010424 | 0.02 |
ENST00000541107.1
ENST00000450218.1 ENST00000326771.2 |
FHL5
|
four and a half LIM domains 5 |
| chr5_-_114505624 | 0.02 |
ENST00000513154.1
|
TRIM36
|
tripartite motif containing 36 |
| chr19_+_41594377 | 0.02 |
ENST00000330436.3
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13 |
| chr1_-_156721502 | 0.02 |
ENST00000357325.5
|
HDGF
|
hepatoma-derived growth factor |
| chr17_-_34329084 | 0.02 |
ENST00000354059.4
ENST00000536149.1 |
CCL15
CCL14
|
chemokine (C-C motif) ligand 15 chemokine (C-C motif) ligand 14 |
| chr14_-_67826486 | 0.02 |
ENST00000555431.1
ENST00000554236.1 ENST00000555474.1 |
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr1_-_150552006 | 0.02 |
ENST00000307940.3
ENST00000369026.2 |
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related) |
| chr11_-_105948129 | 0.02 |
ENST00000526793.1
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr4_-_96470350 | 0.02 |
ENST00000504962.1
ENST00000453304.1 ENST00000506749.1 |
UNC5C
|
unc-5 homolog C (C. elegans) |
| chr7_+_44646162 | 0.02 |
ENST00000439616.2
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr1_+_144989309 | 0.02 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr4_+_110354928 | 0.01 |
ENST00000504968.2
ENST00000399100.2 ENST00000265175.5 |
SEC24B
|
SEC24 family member B |
| chr4_-_65275162 | 0.01 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr16_+_3508063 | 0.01 |
ENST00000576787.1
ENST00000572942.1 ENST00000576916.1 ENST00000575076.1 ENST00000572131.1 |
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr12_-_120966943 | 0.01 |
ENST00000552443.1
ENST00000547736.1 ENST00000445328.2 ENST00000547943.1 ENST00000288532.6 |
COQ5
|
coenzyme Q5 homolog, methyltransferase (S. cerevisiae) |
| chr15_-_66084428 | 0.01 |
ENST00000443035.3
ENST00000431932.2 |
DENND4A
|
DENN/MADD domain containing 4A |
| chr20_+_39657454 | 0.01 |
ENST00000361337.2
|
TOP1
|
topoisomerase (DNA) I |
| chr8_+_110656344 | 0.01 |
ENST00000499579.1
|
RP11-422N16.3
|
Uncharacterized protein |
| chr2_+_183989157 | 0.01 |
ENST00000541912.1
|
NUP35
|
nucleoporin 35kDa |
| chr9_+_110045537 | 0.01 |
ENST00000358015.3
|
RAD23B
|
RAD23 homolog B (S. cerevisiae) |
| chr7_-_130080977 | 0.01 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr16_-_70323422 | 0.01 |
ENST00000261772.8
|
AARS
|
alanyl-tRNA synthetase |
| chr12_-_54758251 | 0.01 |
ENST00000267015.3
ENST00000551809.1 |
GPR84
|
G protein-coupled receptor 84 |
| chr5_+_95997769 | 0.01 |
ENST00000338252.3
ENST00000508830.1 |
CAST
|
calpastatin |
| chr21_+_35736302 | 0.01 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr17_+_39975455 | 0.01 |
ENST00000455106.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr17_-_73179046 | 0.01 |
ENST00000314523.7
ENST00000420826.2 |
SUMO2
|
small ubiquitin-like modifier 2 |
| chr20_-_49547910 | 0.01 |
ENST00000396032.3
|
ADNP
|
activity-dependent neuroprotector homeobox |
| chr8_-_37824442 | 0.01 |
ENST00000345060.3
|
ADRB3
|
adrenoceptor beta 3 |
| chr6_-_74231303 | 0.01 |
ENST00000309268.6
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr2_-_239197201 | 0.01 |
ENST00000254658.3
|
PER2
|
period circadian clock 2 |
| chr12_+_106994905 | 0.01 |
ENST00000357881.4
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr1_+_45212074 | 0.00 |
ENST00000372217.1
|
KIF2C
|
kinesin family member 2C |
| chr9_-_88356789 | 0.00 |
ENST00000357081.3
ENST00000376081.4 ENST00000337006.4 ENST00000376109.3 |
AGTPBP1
|
ATP/GTP binding protein 1 |
| chr1_-_244013384 | 0.00 |
ENST00000366539.1
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr8_+_77318769 | 0.00 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr13_-_33780133 | 0.00 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr8_+_110552831 | 0.00 |
ENST00000530629.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr1_+_65613513 | 0.00 |
ENST00000395334.2
|
AK4
|
adenylate kinase 4 |
| chr5_-_147162263 | 0.00 |
ENST00000333010.6
ENST00000265272.5 |
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr12_-_122017542 | 0.00 |
ENST00000446152.2
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr9_-_114246635 | 0.00 |
ENST00000338205.5
|
KIAA0368
|
KIAA0368 |
| chr22_+_48972118 | 0.00 |
ENST00000358295.5
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
| chr14_-_21489271 | 0.00 |
ENST00000553593.1
|
NDRG2
|
NDRG family member 2 |
| chr7_+_100210133 | 0.00 |
ENST00000393950.2
ENST00000424091.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr9_+_71819927 | 0.00 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HLF_TEF
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.2 | 0.6 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.4 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.7 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.1 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.2 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.6 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.1 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.2 | GO:1902731 | negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.1 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.4 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.2 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.2 | GO:0070487 | monocyte aggregation(GO:0070487) |
| 0.0 | 0.4 | GO:0045187 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.3 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.4 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.6 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 0.5 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.2 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.7 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.1 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |