Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
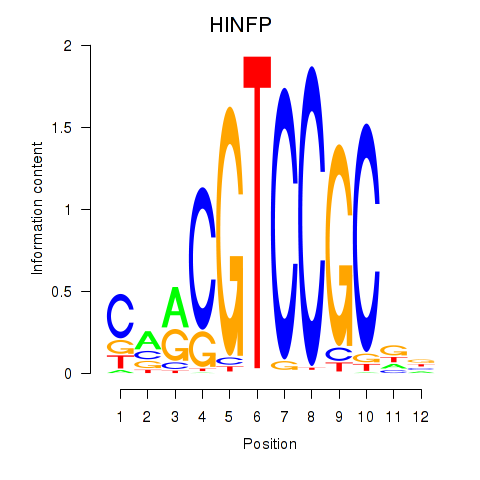
Results for HINFP
Z-value: 0.33
Transcription factors associated with HINFP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HINFP
|
ENSG00000172273.8 | histone H4 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HINFP | hg19_v2_chr11_+_118992269_118992334 | -0.71 | 2.9e-01 | Click! |
Activity profile of HINFP motif
Sorted Z-values of HINFP motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_54963026 | 0.17 |
ENST00000560208.1
ENST00000557792.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr16_-_54962704 | 0.17 |
ENST00000502066.2
ENST00000560912.1 ENST00000558952.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr4_+_166128836 | 0.16 |
ENST00000511305.1
|
KLHL2
|
kelch-like family member 2 |
| chr13_+_111805980 | 0.13 |
ENST00000491775.1
ENST00000466143.1 ENST00000544132.1 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr17_-_73178599 | 0.13 |
ENST00000578238.1
|
SUMO2
|
small ubiquitin-like modifier 2 |
| chrX_-_38186775 | 0.11 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr22_+_29168652 | 0.11 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr12_+_49761147 | 0.11 |
ENST00000549298.1
|
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr6_+_56820018 | 0.11 |
ENST00000370746.3
|
BEND6
|
BEN domain containing 6 |
| chrX_-_38186811 | 0.10 |
ENST00000318842.7
|
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr12_+_70637192 | 0.10 |
ENST00000550160.1
ENST00000551132.1 ENST00000552915.1 ENST00000552483.1 ENST00000550641.1 |
CNOT2
|
CCR4-NOT transcription complex, subunit 2 |
| chr9_+_37120536 | 0.10 |
ENST00000336755.5
ENST00000534928.1 ENST00000322831.6 |
ZCCHC7
|
zinc finger, CCHC domain containing 7 |
| chr12_+_70637494 | 0.09 |
ENST00000548159.1
ENST00000549750.1 ENST00000551043.1 |
CNOT2
|
CCR4-NOT transcription complex, subunit 2 |
| chr5_+_61602236 | 0.09 |
ENST00000514082.1
ENST00000407818.3 |
KIF2A
|
kinesin heavy chain member 2A |
| chr3_-_150481218 | 0.09 |
ENST00000482706.1
|
SIAH2
|
siah E3 ubiquitin protein ligase 2 |
| chr3_-_113415441 | 0.09 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr8_-_102218292 | 0.08 |
ENST00000518336.1
ENST00000520454.1 |
ZNF706
|
zinc finger protein 706 |
| chr16_+_30007524 | 0.08 |
ENST00000567254.1
ENST00000567705.1 |
INO80E
|
INO80 complex subunit E |
| chr6_+_56819773 | 0.08 |
ENST00000370750.2
|
BEND6
|
BEN domain containing 6 |
| chr18_+_268148 | 0.08 |
ENST00000581677.1
|
RP11-705O1.8
|
RP11-705O1.8 |
| chr13_-_36705425 | 0.08 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr2_+_30670209 | 0.08 |
ENST00000497423.1
ENST00000476535.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr2_+_42396574 | 0.08 |
ENST00000401738.3
|
EML4
|
echinoderm microtubule associated protein like 4 |
| chr7_-_140624499 | 0.08 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr13_-_41240717 | 0.07 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr14_+_52118694 | 0.07 |
ENST00000554778.1
|
FRMD6
|
FERM domain containing 6 |
| chr5_+_61601965 | 0.07 |
ENST00000401507.3
|
KIF2A
|
kinesin heavy chain member 2A |
| chr13_+_25875785 | 0.07 |
ENST00000381747.3
|
NUPL1
|
nucleoporin like 1 |
| chr8_+_32406179 | 0.07 |
ENST00000405005.3
|
NRG1
|
neuregulin 1 |
| chr8_-_102217515 | 0.07 |
ENST00000520347.1
ENST00000523922.1 ENST00000520984.1 |
ZNF706
|
zinc finger protein 706 |
| chr18_-_268019 | 0.07 |
ENST00000261600.6
|
THOC1
|
THO complex 1 |
| chrX_+_21392553 | 0.07 |
ENST00000279451.4
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr5_-_141257954 | 0.06 |
ENST00000456271.1
ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1
|
protocadherin 1 |
| chr17_+_30771279 | 0.06 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr16_+_84002234 | 0.06 |
ENST00000305202.4
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr7_+_99006232 | 0.06 |
ENST00000403633.2
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr4_-_18023350 | 0.06 |
ENST00000539056.1
ENST00000382226.5 ENST00000326877.4 |
LCORL
|
ligand dependent nuclear receptor corepressor-like |
| chr5_-_41510725 | 0.06 |
ENST00000328457.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr19_-_39881669 | 0.06 |
ENST00000221266.7
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr19_-_39881777 | 0.06 |
ENST00000595564.1
ENST00000221265.3 |
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr19_-_10530784 | 0.06 |
ENST00000593124.1
|
CDC37
|
cell division cycle 37 |
| chr7_-_17980091 | 0.06 |
ENST00000409389.1
ENST00000409604.1 ENST00000428135.3 |
SNX13
|
sorting nexin 13 |
| chr9_+_100174232 | 0.06 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr13_-_31040060 | 0.06 |
ENST00000326004.4
ENST00000341423.5 |
HMGB1
|
high mobility group box 1 |
| chr12_+_49760639 | 0.06 |
ENST00000549538.1
ENST00000548654.1 ENST00000550643.1 ENST00000548710.1 ENST00000549179.1 ENST00000548377.1 |
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr12_+_69633407 | 0.05 |
ENST00000551516.1
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr10_+_94608245 | 0.05 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr16_+_81069433 | 0.05 |
ENST00000299575.4
|
ATMIN
|
ATM interactor |
| chr1_+_185014647 | 0.05 |
ENST00000367509.4
|
RNF2
|
ring finger protein 2 |
| chr19_+_48112371 | 0.05 |
ENST00000594866.1
|
GLTSCR1
|
glioma tumor suppressor candidate region gene 1 |
| chr2_+_171785824 | 0.05 |
ENST00000452526.2
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chrX_+_21392529 | 0.05 |
ENST00000425654.2
ENST00000543067.1 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr7_-_99006443 | 0.05 |
ENST00000350498.3
|
PDAP1
|
PDGFA associated protein 1 |
| chr14_-_92572894 | 0.05 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
| chr4_+_166128735 | 0.05 |
ENST00000226725.6
|
KLHL2
|
kelch-like family member 2 |
| chr9_+_100174344 | 0.05 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr14_-_99947121 | 0.05 |
ENST00000329331.3
ENST00000436070.2 |
SETD3
|
SET domain containing 3 |
| chr1_+_93913713 | 0.05 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr17_-_73179046 | 0.05 |
ENST00000314523.7
ENST00000420826.2 |
SUMO2
|
small ubiquitin-like modifier 2 |
| chr13_+_111806055 | 0.05 |
ENST00000218789.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr5_-_41510656 | 0.05 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr10_+_124907638 | 0.05 |
ENST00000339992.3
|
HMX2
|
H6 family homeobox 2 |
| chr8_-_11324273 | 0.05 |
ENST00000284486.4
|
FAM167A
|
family with sequence similarity 167, member A |
| chr1_+_61547405 | 0.05 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr10_-_104179682 | 0.05 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chrX_+_154299753 | 0.05 |
ENST00000369459.2
ENST00000369462.1 ENST00000411985.1 ENST00000399042.1 |
BRCC3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr5_-_132073111 | 0.05 |
ENST00000403231.1
|
KIF3A
|
kinesin family member 3A |
| chr5_+_113697983 | 0.05 |
ENST00000264773.3
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr17_-_4269920 | 0.05 |
ENST00000572484.1
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr17_+_7341586 | 0.05 |
ENST00000575235.1
|
FGF11
|
fibroblast growth factor 11 |
| chr2_+_28974489 | 0.05 |
ENST00000455580.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr17_-_6616678 | 0.05 |
ENST00000381074.4
ENST00000293800.6 ENST00000572352.1 ENST00000576323.1 ENST00000573648.1 |
SLC13A5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chrX_+_154299690 | 0.05 |
ENST00000340647.4
ENST00000330045.7 |
BRCC3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr13_+_21714913 | 0.05 |
ENST00000450573.1
ENST00000467636.1 |
SAP18
|
Sin3A-associated protein, 18kDa |
| chr4_+_148402069 | 0.05 |
ENST00000358556.4
ENST00000339690.5 ENST00000511804.1 ENST00000324300.5 |
EDNRA
|
endothelin receptor type A |
| chrX_-_154299501 | 0.05 |
ENST00000369476.3
ENST00000369484.3 |
MTCP1
CMC4
|
mature T-cell proliferation 1 C-x(9)-C motif containing 4 |
| chr8_-_102217796 | 0.05 |
ENST00000519744.1
ENST00000311212.4 ENST00000521272.1 ENST00000519882.1 |
ZNF706
|
zinc finger protein 706 |
| chr2_-_37384175 | 0.05 |
ENST00000411537.2
ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr20_-_30458354 | 0.05 |
ENST00000428829.1
|
DUSP15
|
dual specificity phosphatase 15 |
| chr8_-_100905925 | 0.04 |
ENST00000518171.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr10_-_62761188 | 0.04 |
ENST00000357917.4
|
RHOBTB1
|
Rho-related BTB domain containing 1 |
| chr6_+_35310391 | 0.04 |
ENST00000337400.2
ENST00000311565.4 ENST00000540939.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr2_+_64068116 | 0.04 |
ENST00000480679.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr17_-_66453562 | 0.04 |
ENST00000262139.5
ENST00000546360.1 |
WIPI1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr1_-_197115818 | 0.04 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr2_+_42396472 | 0.04 |
ENST00000318522.5
ENST00000402711.2 |
EML4
|
echinoderm microtubule associated protein like 4 |
| chr15_-_43802769 | 0.04 |
ENST00000263801.3
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr14_+_52118576 | 0.04 |
ENST00000395718.2
ENST00000344768.5 |
FRMD6
|
FERM domain containing 6 |
| chr5_+_65440032 | 0.04 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr3_+_110790590 | 0.04 |
ENST00000485303.1
|
PVRL3
|
poliovirus receptor-related 3 |
| chr13_+_48877895 | 0.04 |
ENST00000267163.4
|
RB1
|
retinoblastoma 1 |
| chr3_+_63897605 | 0.04 |
ENST00000487717.1
|
ATXN7
|
ataxin 7 |
| chr1_-_1509931 | 0.04 |
ENST00000359060.4
|
SSU72
|
SSU72 RNA polymerase II CTD phosphatase homolog (S. cerevisiae) |
| chr7_-_26240357 | 0.04 |
ENST00000354667.4
ENST00000356674.7 |
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr4_+_190861993 | 0.04 |
ENST00000524583.1
ENST00000531991.2 |
FRG1
|
FSHD region gene 1 |
| chr2_+_30670077 | 0.04 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr12_+_102271129 | 0.04 |
ENST00000258534.8
|
DRAM1
|
DNA-damage regulated autophagy modulator 1 |
| chr5_-_127873496 | 0.04 |
ENST00000508989.1
|
FBN2
|
fibrillin 2 |
| chr9_-_94186131 | 0.04 |
ENST00000297689.3
|
NFIL3
|
nuclear factor, interleukin 3 regulated |
| chr13_-_48877795 | 0.04 |
ENST00000436963.1
ENST00000433480.2 |
LINC00441
|
long intergenic non-protein coding RNA 441 |
| chr3_-_129612394 | 0.04 |
ENST00000505616.1
ENST00000426664.2 |
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr17_-_4269768 | 0.04 |
ENST00000396981.2
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr17_+_29158962 | 0.04 |
ENST00000321990.4
|
ATAD5
|
ATPase family, AAA domain containing 5 |
| chr1_+_185014496 | 0.04 |
ENST00000367510.3
|
RNF2
|
ring finger protein 2 |
| chr5_+_61602055 | 0.04 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr12_+_12764773 | 0.04 |
ENST00000228865.2
|
CREBL2
|
cAMP responsive element binding protein-like 2 |
| chr5_-_159546396 | 0.04 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr17_-_42276574 | 0.04 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr10_-_131762105 | 0.04 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr17_+_48450575 | 0.04 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chrX_-_119695279 | 0.04 |
ENST00000336592.6
|
CUL4B
|
cullin 4B |
| chr1_+_61547894 | 0.04 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr13_+_21714653 | 0.04 |
ENST00000382533.4
|
SAP18
|
Sin3A-associated protein, 18kDa |
| chr4_+_492985 | 0.04 |
ENST00000296306.7
ENST00000536264.1 ENST00000310340.5 ENST00000453061.2 ENST00000504346.1 ENST00000503111.1 ENST00000383028.4 ENST00000509768.1 |
PIGG
|
phosphatidylinositol glycan anchor biosynthesis, class G |
| chr2_+_121010413 | 0.04 |
ENST00000404963.3
|
RALB
|
v-ral simian leukemia viral oncogene homolog B |
| chr11_-_46867780 | 0.04 |
ENST00000529230.1
ENST00000415402.1 ENST00000312055.5 |
CKAP5
|
cytoskeleton associated protein 5 |
| chr17_+_12693001 | 0.04 |
ENST00000262444.9
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr17_-_27621125 | 0.03 |
ENST00000579665.1
ENST00000225388.4 |
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr9_-_101017900 | 0.03 |
ENST00000375066.5
|
TBC1D2
|
TBC1 domain family, member 2 |
| chr8_-_30670053 | 0.03 |
ENST00000518564.1
|
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr7_+_94023873 | 0.03 |
ENST00000297268.6
|
COL1A2
|
collagen, type I, alpha 2 |
| chr5_+_142149932 | 0.03 |
ENST00000274498.4
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr15_-_35280426 | 0.03 |
ENST00000559564.1
ENST00000356321.4 |
ZNF770
|
zinc finger protein 770 |
| chr3_+_110790867 | 0.03 |
ENST00000486596.1
ENST00000493615.1 |
PVRL3
|
poliovirus receptor-related 3 |
| chr3_-_49449350 | 0.03 |
ENST00000454011.2
ENST00000445425.1 ENST00000422781.1 |
RHOA
|
ras homolog family member A |
| chr4_+_190861941 | 0.03 |
ENST00000226798.4
|
FRG1
|
FSHD region gene 1 |
| chr2_+_99953816 | 0.03 |
ENST00000289371.6
|
EIF5B
|
eukaryotic translation initiation factor 5B |
| chr2_-_232791038 | 0.03 |
ENST00000295440.2
ENST00000409852.1 |
NPPC
|
natriuretic peptide C |
| chr13_-_31039375 | 0.03 |
ENST00000399494.1
|
HMGB1
|
high mobility group box 1 |
| chr3_-_150481164 | 0.03 |
ENST00000312960.3
|
SIAH2
|
siah E3 ubiquitin protein ligase 2 |
| chr2_+_204193129 | 0.03 |
ENST00000417864.1
|
ABI2
|
abl-interactor 2 |
| chr12_+_90102729 | 0.03 |
ENST00000605386.1
|
LINC00936
|
long intergenic non-protein coding RNA 936 |
| chr9_+_37650945 | 0.03 |
ENST00000377765.3
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr12_+_12510352 | 0.03 |
ENST00000298571.6
|
LOH12CR1
|
loss of heterozygosity, 12, chromosomal region 1 |
| chr20_+_30458431 | 0.03 |
ENST00000375938.4
ENST00000535842.1 ENST00000310998.4 ENST00000375921.2 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr3_-_52719810 | 0.03 |
ENST00000424867.1
ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1
|
polybromo 1 |
| chr1_-_22109682 | 0.03 |
ENST00000400301.1
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chr17_-_28619059 | 0.03 |
ENST00000580709.1
|
BLMH
|
bleomycin hydrolase |
| chr1_+_6845384 | 0.03 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr22_-_22221658 | 0.03 |
ENST00000544786.1
|
MAPK1
|
mitogen-activated protein kinase 1 |
| chr4_-_156298028 | 0.03 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr4_+_123747834 | 0.03 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr12_-_2113583 | 0.03 |
ENST00000397173.4
ENST00000280665.6 |
DCP1B
|
decapping mRNA 1B |
| chr8_+_32405785 | 0.03 |
ENST00000287842.3
|
NRG1
|
neuregulin 1 |
| chr5_+_43121607 | 0.03 |
ENST00000509156.1
ENST00000508259.1 ENST00000306938.4 ENST00000399534.1 |
ZNF131
|
zinc finger protein 131 |
| chr3_-_49377446 | 0.03 |
ENST00000351842.4
ENST00000416417.1 ENST00000415188.1 |
USP4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr11_-_73309112 | 0.03 |
ENST00000450446.2
|
FAM168A
|
family with sequence similarity 168, member A |
| chr14_-_102026643 | 0.03 |
ENST00000555882.1
ENST00000554441.1 ENST00000553729.1 ENST00000557109.1 ENST00000557532.1 ENST00000554694.1 ENST00000554735.1 ENST00000555174.1 ENST00000557661.1 |
DIO3OS
|
DIO3 opposite strand/antisense RNA (head to head) |
| chr5_+_72143988 | 0.03 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr2_+_30670127 | 0.03 |
ENST00000540623.1
ENST00000476038.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr2_+_121010370 | 0.03 |
ENST00000420510.1
|
RALB
|
v-ral simian leukemia viral oncogene homolog B |
| chr1_-_40157345 | 0.03 |
ENST00000372844.3
|
HPCAL4
|
hippocalcin like 4 |
| chr2_+_121010324 | 0.03 |
ENST00000272519.5
|
RALB
|
v-ral simian leukemia viral oncogene homolog B |
| chr2_+_120770645 | 0.03 |
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chrX_-_15353629 | 0.03 |
ENST00000333590.4
ENST00000428964.1 ENST00000542278.1 |
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr17_+_46125685 | 0.03 |
ENST00000579889.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr17_-_1419941 | 0.03 |
ENST00000498390.1
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr12_-_90102594 | 0.03 |
ENST00000428670.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr13_+_25875662 | 0.03 |
ENST00000381736.3
ENST00000463407.1 ENST00000381718.3 |
NUPL1
|
nucleoporin like 1 |
| chr2_-_233352531 | 0.03 |
ENST00000304546.1
|
ECEL1
|
endothelin converting enzyme-like 1 |
| chr3_+_196295482 | 0.03 |
ENST00000440469.1
ENST00000311630.6 |
FBXO45
|
F-box protein 45 |
| chr4_-_6202247 | 0.03 |
ENST00000409021.3
ENST00000409371.3 |
JAKMIP1
|
janus kinase and microtubule interacting protein 1 |
| chr1_-_94703118 | 0.03 |
ENST00000260526.6
ENST00000370217.3 |
ARHGAP29
|
Rho GTPase activating protein 29 |
| chr14_+_64971292 | 0.03 |
ENST00000358738.3
ENST00000394712.2 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr22_+_32871224 | 0.03 |
ENST00000452138.1
ENST00000382058.3 ENST00000397426.1 |
FBXO7
|
F-box protein 7 |
| chr3_-_38691119 | 0.03 |
ENST00000333535.4
ENST00000413689.1 ENST00000443581.1 ENST00000425664.1 ENST00000451551.2 |
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit |
| chr12_+_65672423 | 0.03 |
ENST00000355192.3
ENST00000308259.5 ENST00000540804.1 ENST00000535664.1 ENST00000541189.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr9_-_115095229 | 0.02 |
ENST00000210227.4
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr1_-_1510204 | 0.02 |
ENST00000291386.3
|
SSU72
|
SSU72 RNA polymerase II CTD phosphatase homolog (S. cerevisiae) |
| chr4_+_123747979 | 0.02 |
ENST00000608478.1
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chrX_-_118827113 | 0.02 |
ENST00000394617.2
|
SEPT6
|
septin 6 |
| chr1_+_6845578 | 0.02 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr2_-_73053126 | 0.02 |
ENST00000272427.6
ENST00000410104.1 |
EXOC6B
|
exocyst complex component 6B |
| chr8_+_32406137 | 0.02 |
ENST00000521670.1
|
NRG1
|
neuregulin 1 |
| chr5_-_158526693 | 0.02 |
ENST00000380654.4
|
EBF1
|
early B-cell factor 1 |
| chr4_-_74124502 | 0.02 |
ENST00000358602.4
ENST00000330838.6 ENST00000561029.1 |
ANKRD17
|
ankyrin repeat domain 17 |
| chr13_+_111806083 | 0.02 |
ENST00000375736.4
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr5_+_43121698 | 0.02 |
ENST00000505606.2
ENST00000509634.1 ENST00000509341.1 |
ZNF131
|
zinc finger protein 131 |
| chr2_-_33824336 | 0.02 |
ENST00000431950.1
ENST00000403368.1 ENST00000441530.2 |
FAM98A
|
family with sequence similarity 98, member A |
| chr3_-_49377499 | 0.02 |
ENST00000265560.4
|
USP4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr5_+_142149955 | 0.02 |
ENST00000378004.3
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr9_+_139921916 | 0.02 |
ENST00000314330.2
|
C9orf139
|
chromosome 9 open reading frame 139 |
| chr13_+_21714711 | 0.02 |
ENST00000607003.1
ENST00000492245.1 |
SAP18
|
Sin3A-associated protein, 18kDa |
| chr12_+_12509990 | 0.02 |
ENST00000542728.1
|
LOH12CR1
|
loss of heterozygosity, 12, chromosomal region 1 |
| chr3_-_16555150 | 0.02 |
ENST00000334133.4
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr1_+_6845497 | 0.02 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr13_-_48612067 | 0.02 |
ENST00000543413.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr16_-_54962415 | 0.02 |
ENST00000501177.3
ENST00000559598.2 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr20_+_42086525 | 0.02 |
ENST00000244020.3
|
SRSF6
|
serine/arginine-rich splicing factor 6 |
| chr16_+_4526341 | 0.02 |
ENST00000458134.3
ENST00000219700.6 ENST00000575120.1 ENST00000572812.1 ENST00000574466.1 ENST00000576827.1 ENST00000570445.1 |
HMOX2
|
heme oxygenase (decycling) 2 |
| chr4_-_6202291 | 0.02 |
ENST00000282924.5
|
JAKMIP1
|
janus kinase and microtubule interacting protein 1 |
| chr1_-_200379129 | 0.02 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr2_-_68479614 | 0.02 |
ENST00000234310.3
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chr13_-_77460525 | 0.02 |
ENST00000377474.2
ENST00000317765.2 |
KCTD12
|
potassium channel tetramerization domain containing 12 |
| chr5_-_132073210 | 0.02 |
ENST00000378735.1
ENST00000378746.4 |
KIF3A
|
kinesin family member 3A |
| chr10_+_94352956 | 0.02 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr6_-_111804905 | 0.02 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr2_+_64068074 | 0.02 |
ENST00000394417.2
ENST00000484142.1 ENST00000482668.1 ENST00000467648.2 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr4_-_149363662 | 0.02 |
ENST00000355292.3
ENST00000358102.3 |
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr8_-_100905850 | 0.02 |
ENST00000520271.1
ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C
|
cytochrome c oxidase subunit VIc |
Network of associatons between targets according to the STRING database.
First level regulatory network of HINFP
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:2000426 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.1 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.0 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.0 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.0 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.0 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.0 | GO:0019858 | cytosine metabolic process(GO:0019858) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.0 | GO:0004132 | dCMP deaminase activity(GO:0004132) |