Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
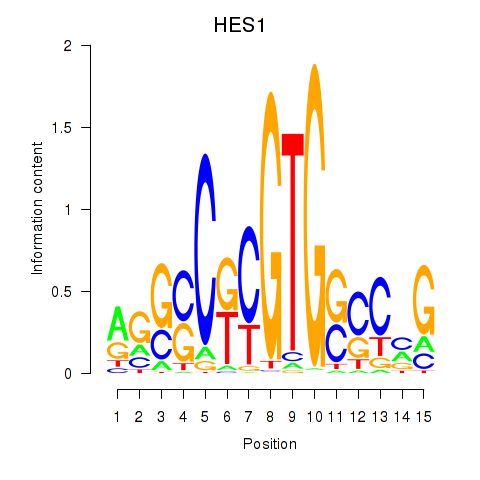
Results for HES1
Z-value: 0.33
Transcription factors associated with HES1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HES1
|
ENSG00000114315.3 | hes family bHLH transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HES1 | hg19_v2_chr3_+_193853927_193853944 | -0.18 | 8.2e-01 | Click! |
Activity profile of HES1 motif
Sorted Z-values of HES1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_77793891 | 0.09 |
ENST00000592957.1
ENST00000585474.1 |
TXNL4A
|
thioredoxin-like 4A |
| chr19_+_54960790 | 0.07 |
ENST00000443957.1
|
LENG8
|
leukocyte receptor cluster (LRC) member 8 |
| chr1_-_9189229 | 0.07 |
ENST00000377411.4
|
GPR157
|
G protein-coupled receptor 157 |
| chr3_+_184081213 | 0.07 |
ENST00000429568.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr3_+_184081137 | 0.06 |
ENST00000443489.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr15_-_44069741 | 0.06 |
ENST00000319359.3
|
ELL3
|
elongation factor RNA polymerase II-like 3 |
| chr11_+_810221 | 0.06 |
ENST00000530398.1
|
RPLP2
|
ribosomal protein, large, P2 |
| chr7_-_558876 | 0.05 |
ENST00000354513.5
ENST00000402802.3 |
PDGFA
|
platelet-derived growth factor alpha polypeptide |
| chr19_-_13030071 | 0.05 |
ENST00000293695.7
|
SYCE2
|
synaptonemal complex central element protein 2 |
| chr14_+_69726864 | 0.05 |
ENST00000448469.3
|
GALNT16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr11_-_47470682 | 0.05 |
ENST00000529341.1
ENST00000352508.3 |
RAPSN
|
receptor-associated protein of the synapse |
| chr1_+_6845578 | 0.05 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr12_+_57854274 | 0.05 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr4_-_10459009 | 0.05 |
ENST00000507515.1
|
ZNF518B
|
zinc finger protein 518B |
| chr2_+_86116396 | 0.05 |
ENST00000455121.3
|
AC105053.4
|
AC105053.4 |
| chr14_+_69726968 | 0.05 |
ENST00000553669.1
|
GALNT16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr19_-_39832563 | 0.05 |
ENST00000599274.1
|
CTC-246B18.10
|
CTC-246B18.10 |
| chr19_+_843314 | 0.05 |
ENST00000544537.2
|
PRTN3
|
proteinase 3 |
| chr3_+_184081175 | 0.04 |
ENST00000452961.1
ENST00000296223.3 |
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr1_-_46152174 | 0.04 |
ENST00000290795.3
ENST00000355105.3 |
GPBP1L1
|
GC-rich promoter binding protein 1-like 1 |
| chr12_-_33049690 | 0.04 |
ENST00000070846.6
ENST00000340811.4 |
PKP2
|
plakophilin 2 |
| chr1_+_6845497 | 0.04 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr8_-_69243711 | 0.04 |
ENST00000512294.3
|
RP11-664D7.4
|
HCG1787533; Uncharacterized protein |
| chr9_-_107754034 | 0.04 |
ENST00000457720.1
|
RP11-217B7.3
|
RP11-217B7.3 |
| chr12_-_122238913 | 0.04 |
ENST00000537157.1
|
AC084018.1
|
AC084018.1 |
| chr17_+_73257945 | 0.04 |
ENST00000579002.1
|
MRPS7
|
mitochondrial ribosomal protein S7 |
| chr2_+_204193149 | 0.03 |
ENST00000422511.2
|
ABI2
|
abl-interactor 2 |
| chr7_-_32931623 | 0.03 |
ENST00000452926.1
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr1_+_840205 | 0.03 |
ENST00000607769.1
|
RP11-54O7.16
|
RP11-54O7.16 |
| chr6_-_150390202 | 0.03 |
ENST00000438272.2
ENST00000367339.2 |
ULBP3
|
UL16 binding protein 3 |
| chr1_+_44445643 | 0.03 |
ENST00000309519.7
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr12_-_129308041 | 0.03 |
ENST00000376740.4
|
SLC15A4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr19_-_14628234 | 0.03 |
ENST00000595139.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr7_-_16460863 | 0.03 |
ENST00000407010.2
ENST00000399310.3 |
ISPD
|
isoprenoid synthase domain containing |
| chr10_+_71561630 | 0.03 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr6_+_37787704 | 0.03 |
ENST00000474522.1
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr16_+_85833274 | 0.03 |
ENST00000568794.1
ENST00000253452.2 ENST00000562336.1 ENST00000561569.1 ENST00000566405.1 |
COX4I1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr1_-_9189144 | 0.03 |
ENST00000414642.2
|
GPR157
|
G protein-coupled receptor 157 |
| chr18_-_33077868 | 0.03 |
ENST00000590757.1
ENST00000592173.1 ENST00000441607.2 ENST00000587450.1 ENST00000589258.1 |
INO80C
RP11-322E11.6
|
INO80 complex subunit C Uncharacterized protein |
| chr3_-_179169181 | 0.03 |
ENST00000497513.1
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr1_+_44445549 | 0.03 |
ENST00000356836.6
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr16_-_71918033 | 0.03 |
ENST00000425432.1
ENST00000313565.6 ENST00000568666.1 ENST00000562797.1 ENST00000564134.1 |
ZNF821
|
zinc finger protein 821 |
| chr8_+_66556936 | 0.03 |
ENST00000262146.4
|
MTFR1
|
mitochondrial fission regulator 1 |
| chr15_-_32695396 | 0.03 |
ENST00000512626.2
ENST00000435655.2 |
GOLGA8K
AC139426.2
|
golgin A8 family, member K Uncharacterized protein; cDNA FLJ52611 |
| chr11_+_86748863 | 0.03 |
ENST00000340353.7
|
TMEM135
|
transmembrane protein 135 |
| chr5_+_612387 | 0.03 |
ENST00000264935.5
ENST00000444221.1 |
CEP72
|
centrosomal protein 72kDa |
| chr4_-_10458982 | 0.03 |
ENST00000326756.3
|
ZNF518B
|
zinc finger protein 518B |
| chr14_-_75330537 | 0.03 |
ENST00000556084.2
ENST00000556489.2 ENST00000445876.1 |
PROX2
|
prospero homeobox 2 |
| chr22_+_24115000 | 0.03 |
ENST00000215743.3
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3) |
| chr13_-_21099935 | 0.03 |
ENST00000298248.7
ENST00000382812.1 |
CRYL1
|
crystallin, lambda 1 |
| chr5_-_178017355 | 0.03 |
ENST00000390654.3
|
COL23A1
|
collagen, type XXIII, alpha 1 |
| chr2_-_130956006 | 0.03 |
ENST00000312988.7
|
TUBA3E
|
tubulin, alpha 3e |
| chr1_-_205290865 | 0.03 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr11_-_47470703 | 0.03 |
ENST00000298854.2
|
RAPSN
|
receptor-associated protein of the synapse |
| chr17_+_29815113 | 0.03 |
ENST00000583755.1
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II) |
| chr1_-_161039647 | 0.03 |
ENST00000368013.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr6_-_13711773 | 0.03 |
ENST00000011619.3
|
RANBP9
|
RAN binding protein 9 |
| chr17_+_74723031 | 0.03 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr2_+_204192942 | 0.03 |
ENST00000295851.5
ENST00000261017.5 |
ABI2
|
abl-interactor 2 |
| chr19_-_56988677 | 0.03 |
ENST00000504904.3
ENST00000292069.6 |
ZNF667
|
zinc finger protein 667 |
| chr22_+_20105259 | 0.03 |
ENST00000416427.1
ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1
|
RAN binding protein 1 |
| chr19_-_13068012 | 0.02 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr12_+_96588368 | 0.02 |
ENST00000547860.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr1_+_94884023 | 0.02 |
ENST00000315713.5
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr14_-_81687197 | 0.02 |
ENST00000553612.1
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chr17_+_79651007 | 0.02 |
ENST00000572392.1
ENST00000577012.1 |
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr7_+_94139105 | 0.02 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1 |
| chr12_+_3069037 | 0.02 |
ENST00000397122.2
|
TEAD4
|
TEA domain family member 4 |
| chr7_-_27239703 | 0.02 |
ENST00000222753.4
|
HOXA13
|
homeobox A13 |
| chr10_-_75634219 | 0.02 |
ENST00000305762.7
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr12_+_131356582 | 0.02 |
ENST00000448750.3
ENST00000541630.1 ENST00000392369.2 ENST00000254675.3 ENST00000535090.1 ENST00000392367.3 |
RAN
|
RAN, member RAS oncogene family |
| chr14_+_74058410 | 0.02 |
ENST00000326303.4
|
ACOT4
|
acyl-CoA thioesterase 4 |
| chr7_-_44365020 | 0.02 |
ENST00000395747.2
ENST00000347193.4 ENST00000346990.4 ENST00000258682.6 ENST00000353625.4 ENST00000421607.1 ENST00000424197.1 ENST00000502837.2 ENST00000350811.3 ENST00000395749.2 |
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta |
| chr21_+_47878757 | 0.02 |
ENST00000400274.1
ENST00000427143.2 ENST00000318711.7 ENST00000457905.3 ENST00000466639.1 ENST00000435722.3 ENST00000417564.2 |
DIP2A
|
DIP2 disco-interacting protein 2 homolog A (Drosophila) |
| chr12_-_124018252 | 0.02 |
ENST00000376874.4
|
RILPL1
|
Rab interacting lysosomal protein-like 1 |
| chr12_+_26111823 | 0.02 |
ENST00000381352.3
ENST00000535907.1 ENST00000405154.2 |
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr7_-_130598059 | 0.02 |
ENST00000432045.2
|
MIR29B1
|
microRNA 29a |
| chr3_+_127348005 | 0.02 |
ENST00000342480.6
|
PODXL2
|
podocalyxin-like 2 |
| chr21_+_35445827 | 0.02 |
ENST00000608209.1
ENST00000381151.3 |
SLC5A3
SLC5A3
|
sodium/myo-inositol cotransporter solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
| chr1_+_28844778 | 0.02 |
ENST00000411533.1
|
RCC1
|
regulator of chromosome condensation 1 |
| chr22_+_29279552 | 0.02 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chr2_-_224702201 | 0.02 |
ENST00000446015.2
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr1_+_110693103 | 0.02 |
ENST00000331565.4
|
SLC6A17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr19_+_34855874 | 0.02 |
ENST00000588991.2
|
GPI
|
glucose-6-phosphate isomerase |
| chr17_+_30813576 | 0.02 |
ENST00000313401.3
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr1_+_41445413 | 0.02 |
ENST00000541520.1
|
CTPS1
|
CTP synthase 1 |
| chr2_-_40006289 | 0.02 |
ENST00000260619.6
ENST00000454352.2 |
THUMPD2
|
THUMP domain containing 2 |
| chrX_-_140271249 | 0.02 |
ENST00000370526.2
|
LDOC1
|
leucine zipper, down-regulated in cancer 1 |
| chr17_+_54671047 | 0.02 |
ENST00000332822.4
|
NOG
|
noggin |
| chr19_+_54960358 | 0.02 |
ENST00000439657.1
ENST00000376514.2 ENST00000376526.4 ENST00000436479.1 |
LENG8
|
leukocyte receptor cluster (LRC) member 8 |
| chr1_-_98386543 | 0.02 |
ENST00000423006.2
ENST00000370192.3 ENST00000306031.5 |
DPYD
|
dihydropyrimidine dehydrogenase |
| chr19_+_34856141 | 0.02 |
ENST00000586425.1
|
GPI
|
glucose-6-phosphate isomerase |
| chr22_-_22090043 | 0.02 |
ENST00000403503.1
|
YPEL1
|
yippee-like 1 (Drosophila) |
| chr1_+_225965518 | 0.02 |
ENST00000304786.7
ENST00000366839.4 ENST00000366838.1 |
SRP9
|
signal recognition particle 9kDa |
| chr8_+_32406179 | 0.02 |
ENST00000405005.3
|
NRG1
|
neuregulin 1 |
| chr17_+_27052892 | 0.02 |
ENST00000579671.1
ENST00000579060.1 |
NEK8
|
NIMA-related kinase 8 |
| chr12_+_96588279 | 0.02 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr18_+_77905794 | 0.02 |
ENST00000587254.1
ENST00000586421.1 |
AC139100.2
|
Uncharacterized protein |
| chr21_+_35445811 | 0.02 |
ENST00000399312.2
|
MRPS6
|
mitochondrial ribosomal protein S6 |
| chr10_-_97050777 | 0.02 |
ENST00000329399.6
|
PDLIM1
|
PDZ and LIM domain 1 |
| chr22_+_46731596 | 0.02 |
ENST00000381019.3
|
TRMU
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr3_-_47619623 | 0.02 |
ENST00000456150.1
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C) |
| chr6_+_43738444 | 0.02 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr2_-_175351744 | 0.02 |
ENST00000295500.4
ENST00000392552.2 ENST00000392551.2 |
GPR155
|
G protein-coupled receptor 155 |
| chr2_+_201390843 | 0.02 |
ENST00000357799.4
ENST00000409203.3 |
SGOL2
|
shugoshin-like 2 (S. pombe) |
| chr2_-_232791038 | 0.02 |
ENST00000295440.2
ENST00000409852.1 |
NPPC
|
natriuretic peptide C |
| chr19_-_48673552 | 0.02 |
ENST00000536218.1
ENST00000596549.1 |
LIG1
|
ligase I, DNA, ATP-dependent |
| chr19_-_14628645 | 0.02 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr11_+_134146635 | 0.02 |
ENST00000431683.2
|
GLB1L3
|
galactosidase, beta 1-like 3 |
| chr1_-_161039753 | 0.02 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr17_+_75137460 | 0.02 |
ENST00000587820.1
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr21_-_44527613 | 0.02 |
ENST00000380276.2
ENST00000398137.1 ENST00000291552.4 |
U2AF1
|
U2 small nuclear RNA auxiliary factor 1 |
| chr20_-_23669590 | 0.02 |
ENST00000217423.3
|
CST4
|
cystatin S |
| chr1_+_179712298 | 0.02 |
ENST00000341785.4
|
FAM163A
|
family with sequence similarity 163, member A |
| chr11_-_71318487 | 0.02 |
ENST00000343767.3
|
AP000867.1
|
AP000867.1 |
| chr18_+_59992527 | 0.02 |
ENST00000586569.1
|
TNFRSF11A
|
tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
| chr1_-_161039456 | 0.02 |
ENST00000368016.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr16_-_28415162 | 0.02 |
ENST00000398944.3
ENST00000398943.3 ENST00000380876.4 |
EIF3CL
|
eukaryotic translation initiation factor 3, subunit C-like |
| chr16_-_72127456 | 0.02 |
ENST00000562153.1
|
TXNL4B
|
thioredoxin-like 4B |
| chr1_-_45806053 | 0.02 |
ENST00000412971.1
ENST00000372098.3 ENST00000372110.3 ENST00000529984.1 ENST00000528332.2 ENST00000372115.3 ENST00000450313.1 |
MUTYH
|
mutY homolog |
| chr18_-_45457478 | 0.02 |
ENST00000402690.2
ENST00000356825.4 |
SMAD2
|
SMAD family member 2 |
| chr18_+_12947981 | 0.02 |
ENST00000262124.11
|
SEH1L
|
SEH1-like (S. cerevisiae) |
| chrX_-_135849484 | 0.02 |
ENST00000370620.1
ENST00000535227.1 |
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr16_-_4401258 | 0.02 |
ENST00000577031.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr6_-_74230741 | 0.02 |
ENST00000316292.9
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr7_-_140340576 | 0.02 |
ENST00000275884.6
ENST00000475837.1 |
DENND2A
|
DENN/MADD domain containing 2A |
| chr7_-_47621229 | 0.02 |
ENST00000434451.1
|
TNS3
|
tensin 3 |
| chr17_+_45726803 | 0.02 |
ENST00000535458.2
ENST00000583648.1 |
KPNB1
|
karyopherin (importin) beta 1 |
| chr2_-_86116093 | 0.02 |
ENST00000377332.3
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr8_-_99954788 | 0.02 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr10_+_75936444 | 0.02 |
ENST00000372734.3
ENST00000541550.1 |
ADK
|
adenosine kinase |
| chr20_-_31124186 | 0.02 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr14_+_57857262 | 0.02 |
ENST00000555166.1
ENST00000556492.1 ENST00000554703.1 |
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr1_-_118472216 | 0.02 |
ENST00000369443.5
|
GDAP2
|
ganglioside induced differentiation associated protein 2 |
| chr16_+_318638 | 0.02 |
ENST00000412541.1
ENST00000435035.1 |
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr16_+_28722809 | 0.02 |
ENST00000566866.1
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C |
| chr6_+_86159821 | 0.02 |
ENST00000369651.3
|
NT5E
|
5'-nucleotidase, ecto (CD73) |
| chrX_-_153095813 | 0.02 |
ENST00000544474.1
|
PDZD4
|
PDZ domain containing 4 |
| chr7_-_98030360 | 0.02 |
ENST00000005260.8
|
BAIAP2L1
|
BAI1-associated protein 2-like 1 |
| chr21_-_38445443 | 0.02 |
ENST00000360525.4
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr2_+_204193129 | 0.02 |
ENST00000417864.1
|
ABI2
|
abl-interactor 2 |
| chr18_+_77794446 | 0.02 |
ENST00000262197.7
|
RBFA
|
ribosome binding factor A (putative) |
| chr19_+_8274204 | 0.02 |
ENST00000561053.1
ENST00000251363.5 ENST00000559450.1 ENST00000559336.1 |
CERS4
|
ceramide synthase 4 |
| chr17_-_31404 | 0.02 |
ENST00000343572.7
|
DOC2B
|
double C2-like domains, beta |
| chr12_+_3068544 | 0.02 |
ENST00000540314.1
ENST00000536826.1 ENST00000359864.2 |
TEAD4
|
TEA domain family member 4 |
| chr14_-_64804814 | 0.02 |
ENST00000554572.1
ENST00000358599.5 |
ESR2
|
estrogen receptor 2 (ER beta) |
| chr22_-_19512893 | 0.02 |
ENST00000403084.1
ENST00000413119.2 |
CLDN5
|
claudin 5 |
| chr1_+_154947126 | 0.02 |
ENST00000368439.1
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr18_+_77794358 | 0.02 |
ENST00000306735.5
|
RBFA
|
ribosome binding factor A (putative) |
| chr5_-_96143602 | 0.02 |
ENST00000443439.2
ENST00000503921.1 ENST00000508227.1 ENST00000507154.1 |
ERAP1
|
endoplasmic reticulum aminopeptidase 1 |
| chr9_-_111696224 | 0.02 |
ENST00000537196.1
|
IKBKAP
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
| chr12_+_133707161 | 0.02 |
ENST00000538918.1
ENST00000540609.1 ENST00000248211.6 ENST00000543271.1 ENST00000536877.1 |
ZNF10
|
zinc finger protein 10 |
| chr11_-_105948129 | 0.02 |
ENST00000526793.1
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr17_-_19265855 | 0.02 |
ENST00000440841.1
ENST00000395615.1 ENST00000461069.2 |
B9D1
|
B9 protein domain 1 |
| chr9_+_101867387 | 0.02 |
ENST00000374990.2
ENST00000552516.1 |
TGFBR1
|
transforming growth factor, beta receptor 1 |
| chr8_-_144679532 | 0.02 |
ENST00000534380.1
ENST00000533494.1 ENST00000531218.1 ENST00000526340.1 ENST00000533204.1 ENST00000532400.1 ENST00000529516.1 ENST00000534377.1 ENST00000531621.1 ENST00000530191.1 ENST00000524900.1 ENST00000526838.1 ENST00000531931.1 ENST00000534475.1 ENST00000442189.2 ENST00000524624.1 ENST00000532596.1 ENST00000529832.1 ENST00000530306.1 ENST00000530545.1 ENST00000525261.1 ENST00000534804.1 ENST00000528303.1 ENST00000528610.1 |
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chrX_-_16888276 | 0.02 |
ENST00000493145.1
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr9_-_88897426 | 0.02 |
ENST00000375991.4
ENST00000326094.4 |
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr14_-_101014435 | 0.02 |
ENST00000554356.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr12_-_107487604 | 0.02 |
ENST00000008527.5
|
CRY1
|
cryptochrome 1 (photolyase-like) |
| chr1_-_53793725 | 0.02 |
ENST00000371454.2
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr2_-_225907150 | 0.02 |
ENST00000258390.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr15_+_30896329 | 0.02 |
ENST00000566740.1
|
GOLGA8H
|
golgin A8 family, member H |
| chr1_-_153949751 | 0.02 |
ENST00000428469.1
|
JTB
|
jumping translocation breakpoint |
| chr2_-_177502254 | 0.02 |
ENST00000339037.3
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr12_+_7053228 | 0.02 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr1_-_45805607 | 0.02 |
ENST00000372104.1
ENST00000448481.1 ENST00000483127.1 ENST00000528013.2 ENST00000456914.2 |
MUTYH
|
mutY homolog |
| chr2_+_27237615 | 0.02 |
ENST00000458529.1
ENST00000402218.1 |
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr11_+_64058758 | 0.02 |
ENST00000538767.1
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr3_+_10857885 | 0.02 |
ENST00000254488.2
ENST00000454147.1 |
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr12_-_124457257 | 0.02 |
ENST00000545891.1
|
CCDC92
|
coiled-coil domain containing 92 |
| chr10_+_120789223 | 0.02 |
ENST00000425699.1
|
NANOS1
|
nanos homolog 1 (Drosophila) |
| chr14_+_93260642 | 0.02 |
ENST00000355976.2
|
GOLGA5
|
golgin A5 |
| chr17_-_7297519 | 0.02 |
ENST00000576362.1
ENST00000571078.1 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chrX_-_51239425 | 0.02 |
ENST00000375992.3
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr17_+_77681075 | 0.02 |
ENST00000397549.2
|
CTD-2116F7.1
|
CTD-2116F7.1 |
| chr17_-_17109579 | 0.02 |
ENST00000321560.3
|
PLD6
|
phospholipase D family, member 6 |
| chr2_+_219433588 | 0.02 |
ENST00000295701.5
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr11_+_64058820 | 0.02 |
ENST00000422670.2
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr15_+_84116106 | 0.02 |
ENST00000535412.1
ENST00000324537.5 |
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr5_-_149380195 | 0.02 |
ENST00000515406.2
|
TIGD6
|
tigger transposable element derived 6 |
| chr5_-_1344999 | 0.02 |
ENST00000320927.6
|
CLPTM1L
|
CLPTM1-like |
| chr14_-_60043524 | 0.02 |
ENST00000537690.2
ENST00000281581.4 |
CCDC175
|
coiled-coil domain containing 175 |
| chr20_-_50385138 | 0.02 |
ENST00000338821.5
|
ATP9A
|
ATPase, class II, type 9A |
| chr16_+_21312170 | 0.02 |
ENST00000338573.5
ENST00000561968.1 |
CRYM-AS1
|
CRYM antisense RNA 1 |
| chr12_+_49961990 | 0.02 |
ENST00000551063.1
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr2_-_136743039 | 0.02 |
ENST00000537273.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr8_-_1922789 | 0.02 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr22_+_44351301 | 0.02 |
ENST00000350028.4
|
SAMM50
|
SAMM50 sorting and assembly machinery component |
| chrX_-_47004878 | 0.02 |
ENST00000377811.3
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr21_-_10990830 | 0.02 |
ENST00000361285.4
ENST00000342420.5 ENST00000328758.5 |
TPTE
|
transmembrane phosphatase with tensin homology |
| chr12_-_48213735 | 0.02 |
ENST00000417902.1
ENST00000417107.1 |
HDAC7
|
histone deacetylase 7 |
| chr16_-_71496112 | 0.02 |
ENST00000393539.2
ENST00000417828.1 ENST00000565718.1 ENST00000497160.1 ENST00000428724.2 |
ZNF23
|
zinc finger protein 23 |
| chr1_+_11751748 | 0.02 |
ENST00000294485.5
|
DRAXIN
|
dorsal inhibitory axon guidance protein |
| chr7_-_44365216 | 0.02 |
ENST00000358707.3
ENST00000457475.1 ENST00000440254.2 |
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta |
| chr14_-_53162361 | 0.02 |
ENST00000395686.3
|
ERO1L
|
ERO1-like (S. cerevisiae) |
| chr17_-_79849438 | 0.02 |
ENST00000331204.4
ENST00000505490.2 |
ALYREF
|
Aly/REF export factor |
| chr1_-_31196427 | 0.02 |
ENST00000373765.4
|
MATN1
|
matrilin 1, cartilage matrix protein |
| chr1_+_64936428 | 0.02 |
ENST00000371073.2
ENST00000290039.5 |
CACHD1
|
cache domain containing 1 |
| chrX_-_154033661 | 0.02 |
ENST00000393531.1
|
MPP1
|
membrane protein, palmitoylated 1, 55kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of HES1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.0 | GO:0071810 | regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.0 | 0.0 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.0 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.0 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.0 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.0 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.0 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.0 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.0 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |