Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
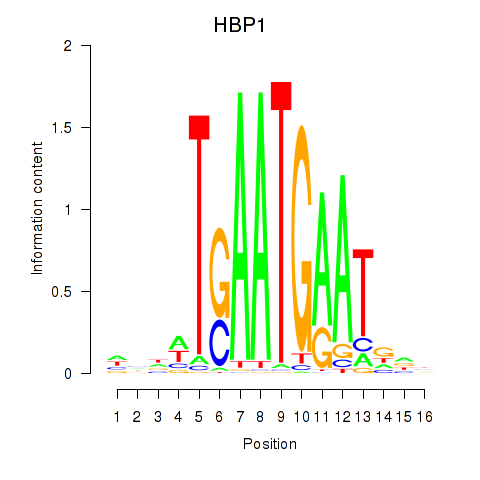
Results for HBP1
Z-value: 0.82
Transcription factors associated with HBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HBP1
|
ENSG00000105856.9 | HMG-box transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HBP1 | hg19_v2_chr7_+_106809406_106809460 | -0.91 | 9.4e-02 | Click! |
Activity profile of HBP1 motif
Sorted Z-values of HBP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_54267147 | 0.44 |
ENST00000558866.1
ENST00000558920.1 |
RP11-643A5.2
|
RP11-643A5.2 |
| chr12_-_11214893 | 0.43 |
ENST00000533467.1
|
TAS2R46
|
taste receptor, type 2, member 46 |
| chr16_+_77246337 | 0.42 |
ENST00000563157.1
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr19_+_49055332 | 0.39 |
ENST00000201586.2
|
SULT2B1
|
sulfotransferase family, cytosolic, 2B, member 1 |
| chr19_-_56092187 | 0.38 |
ENST00000325421.4
ENST00000592239.1 |
ZNF579
|
zinc finger protein 579 |
| chr11_+_63742050 | 0.32 |
ENST00000314133.3
ENST00000535431.1 |
COX8A
AP000721.4
|
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr22_+_24236191 | 0.32 |
ENST00000215754.7
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor) |
| chr6_+_135818979 | 0.31 |
ENST00000421378.2
ENST00000579057.1 ENST00000436554.1 ENST00000438618.2 |
LINC00271
|
long intergenic non-protein coding RNA 271 |
| chr2_+_87808725 | 0.31 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr6_-_44233361 | 0.30 |
ENST00000275015.5
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr5_+_150040403 | 0.30 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr16_+_57139933 | 0.30 |
ENST00000566259.1
|
CPNE2
|
copine II |
| chr6_+_135818907 | 0.29 |
ENST00000579339.1
ENST00000580741.1 |
LINC00271
|
long intergenic non-protein coding RNA 271 |
| chr19_-_50464338 | 0.26 |
ENST00000426971.2
ENST00000447370.2 |
SIGLEC11
|
sialic acid binding Ig-like lectin 11 |
| chrX_+_18725758 | 0.26 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr11_+_66824346 | 0.25 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr18_-_56985776 | 0.24 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr1_-_24127256 | 0.23 |
ENST00000418277.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr7_+_65540853 | 0.23 |
ENST00000380839.4
ENST00000395332.3 ENST00000362000.5 ENST00000395331.3 |
ASL
|
argininosuccinate lyase |
| chr12_-_89746264 | 0.23 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr22_+_25615489 | 0.23 |
ENST00000398215.2
|
CRYBB2
|
crystallin, beta B2 |
| chr2_+_242498135 | 0.22 |
ENST00000318407.3
|
BOK
|
BCL2-related ovarian killer |
| chr8_-_27457494 | 0.22 |
ENST00000521770.1
|
CLU
|
clusterin |
| chr7_+_65540780 | 0.21 |
ENST00000304874.9
|
ASL
|
argininosuccinate lyase |
| chr1_-_27693349 | 0.21 |
ENST00000374040.3
ENST00000357582.2 ENST00000493901.1 |
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr5_-_66942617 | 0.21 |
ENST00000507298.1
|
RP11-83M16.5
|
RP11-83M16.5 |
| chr7_+_74379083 | 0.20 |
ENST00000361825.7
|
GATSL1
|
GATS protein-like 1 |
| chr11_-_68611721 | 0.20 |
ENST00000561996.1
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver) |
| chr19_+_17830051 | 0.20 |
ENST00000594625.1
ENST00000324096.4 ENST00000600186.1 ENST00000597735.1 |
MAP1S
|
microtubule-associated protein 1S |
| chr11_+_122709200 | 0.20 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr11_-_66725837 | 0.20 |
ENST00000393958.2
ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC
|
pyruvate carboxylase |
| chr3_+_37440653 | 0.19 |
ENST00000328376.5
ENST00000452017.2 |
C3orf35
|
chromosome 3 open reading frame 35 |
| chr14_+_32963433 | 0.19 |
ENST00000554410.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr8_-_69243711 | 0.19 |
ENST00000512294.3
|
RP11-664D7.4
|
HCG1787533; Uncharacterized protein |
| chr1_-_16563641 | 0.19 |
ENST00000375599.3
|
RSG1
|
REM2 and RAB-like small GTPase 1 |
| chr17_+_33895090 | 0.19 |
ENST00000592381.1
|
RP11-1094M14.11
|
RP11-1094M14.11 |
| chr14_+_78266436 | 0.18 |
ENST00000557501.1
ENST00000341211.5 |
ADCK1
|
aarF domain containing kinase 1 |
| chr6_-_41909466 | 0.18 |
ENST00000414200.2
|
CCND3
|
cyclin D3 |
| chr1_-_24126892 | 0.17 |
ENST00000374497.3
ENST00000425913.1 |
GALE
|
UDP-galactose-4-epimerase |
| chr2_-_239148599 | 0.17 |
ENST00000409182.1
ENST00000409002.3 ENST00000450098.1 ENST00000409356.1 ENST00000409160.3 ENST00000409574.1 ENST00000272937.5 |
HES6
|
hes family bHLH transcription factor 6 |
| chr12_+_79357815 | 0.17 |
ENST00000547046.1
|
SYT1
|
synaptotagmin I |
| chr19_-_39390440 | 0.17 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr1_-_25291475 | 0.17 |
ENST00000338888.3
ENST00000399916.1 |
RUNX3
|
runt-related transcription factor 3 |
| chr3_-_178103144 | 0.16 |
ENST00000417383.1
ENST00000418585.1 ENST00000411727.1 ENST00000439810.1 |
RP11-33A14.1
|
RP11-33A14.1 |
| chr19_-_3500635 | 0.16 |
ENST00000250937.3
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase |
| chr19_-_38806540 | 0.16 |
ENST00000592694.1
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr19_-_39390350 | 0.15 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chrX_+_54834159 | 0.15 |
ENST00000375053.2
ENST00000347546.4 ENST00000375062.4 |
MAGED2
|
melanoma antigen family D, 2 |
| chr20_-_33872548 | 0.15 |
ENST00000374443.3
|
EIF6
|
eukaryotic translation initiation factor 6 |
| chr14_-_80677815 | 0.15 |
ENST00000557125.1
ENST00000555750.1 |
DIO2
|
deiodinase, iodothyronine, type II |
| chr17_-_27405875 | 0.14 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr17_+_75369400 | 0.14 |
ENST00000590059.1
|
SEPT9
|
septin 9 |
| chr3_+_38537960 | 0.14 |
ENST00000453767.1
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr17_+_36584662 | 0.14 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr2_-_220264703 | 0.14 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr17_+_4853442 | 0.14 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr6_-_41909561 | 0.14 |
ENST00000372991.4
|
CCND3
|
cyclin D3 |
| chr1_+_145209092 | 0.14 |
ENST00000362074.6
ENST00000344859.3 |
NOTCH2NL
|
notch 2 N-terminal like |
| chr18_+_18943554 | 0.14 |
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr19_-_38806560 | 0.14 |
ENST00000591755.1
ENST00000337679.8 ENST00000339413.6 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr1_-_17338267 | 0.14 |
ENST00000326735.8
|
ATP13A2
|
ATPase type 13A2 |
| chr12_-_58027138 | 0.14 |
ENST00000341156.4
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr8_+_104152922 | 0.14 |
ENST00000309982.5
ENST00000438105.2 ENST00000297574.6 |
BAALC
|
brain and acute leukemia, cytoplasmic |
| chr22_+_39745930 | 0.13 |
ENST00000318801.4
ENST00000216155.7 ENST00000406293.3 ENST00000328933.5 |
SYNGR1
|
synaptogyrin 1 |
| chrX_+_54834004 | 0.13 |
ENST00000375068.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr3_+_72937182 | 0.13 |
ENST00000389617.4
|
GXYLT2
|
glucoside xylosyltransferase 2 |
| chr16_+_30953696 | 0.13 |
ENST00000566320.2
ENST00000565939.1 |
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr20_+_10415931 | 0.13 |
ENST00000334534.5
|
SLX4IP
|
SLX4 interacting protein |
| chr9_-_25678856 | 0.12 |
ENST00000358022.3
|
TUSC1
|
tumor suppressor candidate 1 |
| chr19_+_17326521 | 0.12 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr3_-_127455200 | 0.12 |
ENST00000398101.3
|
MGLL
|
monoglyceride lipase |
| chr8_+_66619277 | 0.12 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr11_+_66824276 | 0.12 |
ENST00000308831.2
|
RHOD
|
ras homolog family member D |
| chr9_-_128246769 | 0.12 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr6_-_30640811 | 0.12 |
ENST00000376442.3
|
DHX16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr19_-_45927622 | 0.12 |
ENST00000300853.3
ENST00000589165.1 |
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr15_+_75074385 | 0.12 |
ENST00000220003.9
|
CSK
|
c-src tyrosine kinase |
| chr19_-_42806919 | 0.11 |
ENST00000595530.1
ENST00000538771.1 ENST00000601865.1 |
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr14_-_94759595 | 0.11 |
ENST00000261994.4
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr7_+_99070527 | 0.11 |
ENST00000379724.3
|
ZNF789
|
zinc finger protein 789 |
| chr12_-_121972556 | 0.11 |
ENST00000545022.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr14_-_94759361 | 0.11 |
ENST00000393096.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr11_-_62457371 | 0.11 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr11_+_94277017 | 0.11 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr3_+_46616017 | 0.11 |
ENST00000542931.1
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr14_+_78266408 | 0.11 |
ENST00000238561.5
|
ADCK1
|
aarF domain containing kinase 1 |
| chr7_+_135611542 | 0.11 |
ENST00000416501.1
|
AC015987.2
|
AC015987.2 |
| chr14_-_94759408 | 0.11 |
ENST00000554723.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr16_+_31366455 | 0.11 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr19_+_1040096 | 0.10 |
ENST00000263094.6
ENST00000524850.1 |
ABCA7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr19_-_38806390 | 0.10 |
ENST00000589247.1
ENST00000329420.8 ENST00000591784.1 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr22_-_30956746 | 0.10 |
ENST00000437282.1
ENST00000447224.1 ENST00000427899.1 ENST00000406955.1 ENST00000452827.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr2_+_65216462 | 0.10 |
ENST00000234256.3
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr19_-_45927097 | 0.10 |
ENST00000340192.7
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr1_+_158149737 | 0.10 |
ENST00000368171.3
|
CD1D
|
CD1d molecule |
| chr19_-_42806723 | 0.10 |
ENST00000262890.3
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr19_-_42806444 | 0.10 |
ENST00000594989.1
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr20_+_13765596 | 0.10 |
ENST00000378106.5
ENST00000463598.1 |
NDUFAF5
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 5 |
| chr7_-_56101826 | 0.10 |
ENST00000421626.1
|
PSPH
|
phosphoserine phosphatase |
| chr6_-_41168920 | 0.10 |
ENST00000483722.1
|
TREML2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr15_-_86338134 | 0.10 |
ENST00000337975.5
|
KLHL25
|
kelch-like family member 25 |
| chr12_-_53320245 | 0.09 |
ENST00000552150.1
|
KRT8
|
keratin 8 |
| chr1_-_31769595 | 0.09 |
ENST00000263694.4
|
SNRNP40
|
small nuclear ribonucleoprotein 40kDa (U5) |
| chr11_+_69061594 | 0.09 |
ENST00000441339.2
ENST00000308946.3 ENST00000535407.1 |
MYEOV
|
myeloma overexpressed |
| chr1_+_25598872 | 0.09 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen |
| chr16_+_69140122 | 0.09 |
ENST00000219322.3
|
HAS3
|
hyaluronan synthase 3 |
| chr11_+_22689648 | 0.09 |
ENST00000278187.3
|
GAS2
|
growth arrest-specific 2 |
| chr20_+_25176318 | 0.09 |
ENST00000354989.5
ENST00000360031.2 ENST00000376652.4 ENST00000439162.1 ENST00000433417.1 ENST00000417467.1 ENST00000433259.2 ENST00000427553.1 ENST00000435520.1 ENST00000418890.1 |
ENTPD6
|
ectonucleoside triphosphate diphosphohydrolase 6 (putative) |
| chr7_+_39125365 | 0.09 |
ENST00000559001.1
ENST00000464276.2 |
POU6F2
|
POU class 6 homeobox 2 |
| chr1_+_12185949 | 0.09 |
ENST00000413146.2
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8 |
| chr19_+_39390320 | 0.09 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr4_+_174818390 | 0.09 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr3_+_51575596 | 0.09 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr16_+_57769635 | 0.08 |
ENST00000379661.3
ENST00000562592.1 ENST00000566726.1 |
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr17_-_41856305 | 0.08 |
ENST00000397937.2
ENST00000226004.3 |
DUSP3
|
dual specificity phosphatase 3 |
| chr19_+_34856141 | 0.08 |
ENST00000586425.1
|
GPI
|
glucose-6-phosphate isomerase |
| chr6_+_30614779 | 0.08 |
ENST00000293604.6
ENST00000376473.5 |
C6orf136
|
chromosome 6 open reading frame 136 |
| chr5_+_140762268 | 0.08 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr9_+_86237963 | 0.08 |
ENST00000277124.8
|
IDNK
|
idnK, gluconokinase homolog (E. coli) |
| chr13_-_46626820 | 0.08 |
ENST00000428921.1
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr2_-_157189180 | 0.08 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr3_+_184279566 | 0.08 |
ENST00000330394.2
|
EPHB3
|
EPH receptor B3 |
| chr15_+_75074410 | 0.08 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr7_-_149158187 | 0.08 |
ENST00000247930.4
|
ZNF777
|
zinc finger protein 777 |
| chr3_+_184032419 | 0.08 |
ENST00000352767.3
ENST00000427141.2 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_+_202172848 | 0.08 |
ENST00000255432.7
|
LGR6
|
leucine-rich repeat containing G protein-coupled receptor 6 |
| chr1_+_44435646 | 0.08 |
ENST00000255108.3
ENST00000412950.2 ENST00000396758.2 |
DPH2
|
DPH2 homolog (S. cerevisiae) |
| chr3_+_133524459 | 0.08 |
ENST00000484684.1
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chr15_+_43477580 | 0.08 |
ENST00000356633.5
|
CCNDBP1
|
cyclin D-type binding-protein 1 |
| chr4_-_69083720 | 0.07 |
ENST00000432593.3
|
TMPRSS11BNL
|
TMPRSS11B N-terminal like |
| chr3_+_140770183 | 0.07 |
ENST00000310546.2
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr17_+_8152590 | 0.07 |
ENST00000584044.1
ENST00000314666.6 ENST00000545834.1 ENST00000581242.1 |
PFAS
|
phosphoribosylformylglycinamidine synthase |
| chr22_+_41968007 | 0.07 |
ENST00000460790.1
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr17_-_36904437 | 0.07 |
ENST00000585100.1
ENST00000360797.2 ENST00000578109.1 ENST00000579882.1 |
PCGF2
|
polycomb group ring finger 2 |
| chr12_-_89746173 | 0.07 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr3_+_185000729 | 0.07 |
ENST00000448876.1
ENST00000446828.1 ENST00000447637.1 ENST00000424227.1 ENST00000454237.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr5_+_125758813 | 0.07 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr5_+_149737202 | 0.07 |
ENST00000451292.1
ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1
|
Treacher Collins-Franceschetti syndrome 1 |
| chr10_+_104263743 | 0.07 |
ENST00000369902.3
ENST00000369899.2 ENST00000423559.2 |
SUFU
|
suppressor of fused homolog (Drosophila) |
| chr19_+_34856078 | 0.07 |
ENST00000589399.1
ENST00000589640.1 ENST00000591204.1 |
GPI
|
glucose-6-phosphate isomerase |
| chrX_-_23926004 | 0.07 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr15_+_36871983 | 0.07 |
ENST00000437989.2
ENST00000569302.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr8_-_8751068 | 0.07 |
ENST00000276282.6
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr20_-_1373682 | 0.07 |
ENST00000381724.3
|
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr19_+_34855874 | 0.07 |
ENST00000588991.2
|
GPI
|
glucose-6-phosphate isomerase |
| chr16_-_745946 | 0.06 |
ENST00000562563.1
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr15_+_34261089 | 0.06 |
ENST00000383263.5
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr8_-_11725549 | 0.06 |
ENST00000505496.2
ENST00000534636.1 ENST00000524500.1 ENST00000345125.3 ENST00000453527.2 ENST00000527215.2 ENST00000532392.1 ENST00000533455.1 ENST00000534510.1 ENST00000530640.2 ENST00000531089.1 ENST00000532656.2 ENST00000531502.1 ENST00000434271.1 ENST00000353047.6 |
CTSB
|
cathepsin B |
| chr12_-_58146128 | 0.06 |
ENST00000551800.1
ENST00000549606.1 ENST00000257904.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr7_+_6617039 | 0.06 |
ENST00000405731.3
ENST00000396713.2 ENST00000396707.2 ENST00000335965.6 ENST00000396709.1 ENST00000483589.1 ENST00000396706.2 |
ZDHHC4
|
zinc finger, DHHC-type containing 4 |
| chr12_+_128399965 | 0.06 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr10_+_6130966 | 0.06 |
ENST00000437845.2
ENST00000432931.1 |
RBM17
|
RNA binding motif protein 17 |
| chr16_+_11762270 | 0.06 |
ENST00000329565.5
|
SNN
|
stannin |
| chr20_-_33543546 | 0.06 |
ENST00000216951.2
|
GSS
|
glutathione synthetase |
| chr13_+_88325498 | 0.06 |
ENST00000400028.3
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr12_-_122018859 | 0.06 |
ENST00000536437.1
ENST00000377071.4 ENST00000538046.2 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr15_+_72410660 | 0.06 |
ENST00000564082.1
|
SENP8
|
SUMO/sentrin specific peptidase family member 8 |
| chr2_-_74374995 | 0.06 |
ENST00000295326.4
|
BOLA3
|
bolA family member 3 |
| chr4_-_159094194 | 0.06 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr20_-_33872518 | 0.06 |
ENST00000374436.3
|
EIF6
|
eukaryotic translation initiation factor 6 |
| chr19_+_34855925 | 0.06 |
ENST00000590375.1
ENST00000356487.5 |
GPI
|
glucose-6-phosphate isomerase |
| chr1_-_36863481 | 0.06 |
ENST00000315732.2
|
LSM10
|
LSM10, U7 small nuclear RNA associated |
| chr10_+_104180580 | 0.06 |
ENST00000425536.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr12_+_128399917 | 0.05 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr14_-_80677613 | 0.05 |
ENST00000556811.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr9_+_133589333 | 0.05 |
ENST00000372348.2
ENST00000393293.4 |
ABL1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr3_+_158519654 | 0.05 |
ENST00000415822.2
ENST00000392813.4 ENST00000264266.8 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr22_-_18256742 | 0.05 |
ENST00000317361.7
|
BID
|
BH3 interacting domain death agonist |
| chr22_-_22337146 | 0.05 |
ENST00000398793.2
|
TOP3B
|
topoisomerase (DNA) III beta |
| chr15_+_57884117 | 0.05 |
ENST00000267853.5
|
MYZAP
|
myocardial zonula adherens protein |
| chr5_-_16465901 | 0.05 |
ENST00000308683.2
|
ZNF622
|
zinc finger protein 622 |
| chr3_+_126423045 | 0.05 |
ENST00000290913.3
ENST00000508789.1 |
CHCHD6
|
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chr16_-_30905584 | 0.05 |
ENST00000380317.4
|
BCL7C
|
B-cell CLL/lymphoma 7C |
| chr11_+_57435441 | 0.05 |
ENST00000528177.1
|
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr16_-_3661578 | 0.05 |
ENST00000294008.3
|
SLX4
|
SLX4 structure-specific endonuclease subunit |
| chr2_-_197226875 | 0.05 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr12_+_1099675 | 0.05 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr2_+_235887329 | 0.05 |
ENST00000409212.1
ENST00000344528.4 ENST00000444916.1 |
SH3BP4
|
SH3-domain binding protein 4 |
| chr7_-_83278322 | 0.05 |
ENST00000307792.3
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr8_-_64080945 | 0.05 |
ENST00000603538.1
|
YTHDF3-AS1
|
YTHDF3 antisense RNA 1 (head to head) |
| chr19_+_41698927 | 0.05 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr8_-_116504448 | 0.05 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr9_-_35563896 | 0.05 |
ENST00000399742.2
|
FAM166B
|
family with sequence similarity 166, member B |
| chr17_+_6918093 | 0.05 |
ENST00000439424.2
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr16_-_28223166 | 0.05 |
ENST00000304658.5
|
XPO6
|
exportin 6 |
| chr4_+_153457404 | 0.04 |
ENST00000604157.1
ENST00000594836.1 |
MIR4453
|
microRNA 4453 |
| chr8_-_82359662 | 0.04 |
ENST00000519260.1
ENST00000256103.2 |
PMP2
|
peripheral myelin protein 2 |
| chr8_-_27850180 | 0.04 |
ENST00000380385.2
ENST00000301906.4 ENST00000354914.3 |
SCARA5
|
scavenger receptor class A, member 5 (putative) |
| chr19_+_19144384 | 0.04 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr2_+_29341037 | 0.04 |
ENST00000449202.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr17_+_68071389 | 0.04 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr22_-_22337154 | 0.04 |
ENST00000413067.2
ENST00000437929.1 ENST00000456075.1 ENST00000434517.1 ENST00000424393.1 ENST00000449704.1 ENST00000437103.1 |
TOP3B
|
topoisomerase (DNA) III beta |
| chr6_+_30297306 | 0.04 |
ENST00000420746.1
ENST00000513556.1 |
TRIM39
TRIM39-RPP21
|
tripartite motif containing 39 TRIM39-RPP21 readthrough |
| chrX_+_51636629 | 0.04 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr19_+_39390587 | 0.04 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr3_+_111805262 | 0.04 |
ENST00000484828.1
|
C3orf52
|
chromosome 3 open reading frame 52 |
| chr9_+_78505554 | 0.04 |
ENST00000545128.1
|
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr10_-_120101752 | 0.04 |
ENST00000369170.4
|
FAM204A
|
family with sequence similarity 204, member A |
| chr14_+_102430855 | 0.04 |
ENST00000360184.4
|
DYNC1H1
|
dynein, cytoplasmic 1, heavy chain 1 |
| chr11_+_57435219 | 0.04 |
ENST00000527985.1
ENST00000287169.3 |
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HBP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.1 | 0.3 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.1 | 0.4 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.3 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.1 | 0.3 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.3 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.2 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:1905166 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.0 | 0.2 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.3 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.3 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.1 | GO:1901076 | positive regulation of engulfment of apoptotic cell(GO:1901076) |
| 0.0 | 0.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0032679 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.2 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.2 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.1 | GO:1903314 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.0 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 0.3 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.1 | 0.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.4 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 0.3 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.1 | 0.3 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.4 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.1 | GO:0015186 | L-cystine transmembrane transporter activity(GO:0015184) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |