Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for GUAGUGU
Z-value: 0.28
miRNA associated with seed GUAGUGU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-142-3p.1
|
MIMAT0000434 |
Activity profile of GUAGUGU motif
Sorted Z-values of GUAGUGU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_86948245 | 0.07 |
ENST00000439940.2
ENST00000604011.1 |
CHMP3
RNF103-CHMP3
|
charged multivesicular body protein 3 RNF103-CHMP3 readthrough |
| chr19_-_18314836 | 0.07 |
ENST00000464076.3
ENST00000222256.4 |
RAB3A
|
RAB3A, member RAS oncogene family |
| chr11_+_9406169 | 0.07 |
ENST00000379719.3
ENST00000527431.1 |
IPO7
|
importin 7 |
| chr2_+_153191706 | 0.06 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr1_+_28099683 | 0.06 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chrX_-_135056216 | 0.06 |
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr2_-_11484710 | 0.06 |
ENST00000315872.6
|
ROCK2
|
Rho-associated, coiled-coil containing protein kinase 2 |
| chr4_+_113066552 | 0.06 |
ENST00000309733.5
|
C4orf32
|
chromosome 4 open reading frame 32 |
| chr3_+_20081515 | 0.06 |
ENST00000263754.4
|
KAT2B
|
K(lysine) acetyltransferase 2B |
| chr17_+_30771279 | 0.05 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr1_-_197169672 | 0.05 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr14_-_31495569 | 0.05 |
ENST00000357479.5
ENST00000355683.5 |
STRN3
|
striatin, calmodulin binding protein 3 |
| chr17_-_8534067 | 0.05 |
ENST00000360416.3
ENST00000269243.4 |
MYH10
|
myosin, heavy chain 10, non-muscle |
| chr15_+_85523671 | 0.05 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chr1_+_40627038 | 0.05 |
ENST00000372771.4
|
RLF
|
rearranged L-myc fusion |
| chr1_+_25071848 | 0.05 |
ENST00000374379.4
|
CLIC4
|
chloride intracellular channel 4 |
| chr21_+_30671189 | 0.04 |
ENST00000286800.3
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr12_-_44200052 | 0.04 |
ENST00000548315.1
ENST00000552521.1 ENST00000546662.1 ENST00000548403.1 ENST00000546506.1 |
TWF1
|
twinfilin actin-binding protein 1 |
| chr1_+_24069642 | 0.04 |
ENST00000418390.2
|
TCEB3
|
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A) |
| chr11_-_77532050 | 0.04 |
ENST00000308488.6
|
RSF1
|
remodeling and spacing factor 1 |
| chr16_-_18937726 | 0.04 |
ENST00000389467.3
ENST00000446231.2 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr4_+_6642404 | 0.04 |
ENST00000507420.1
ENST00000382581.4 |
MRFAP1
|
Morf4 family associated protein 1 |
| chr1_+_11072696 | 0.04 |
ENST00000240185.3
ENST00000476201.1 |
TARDBP
|
TAR DNA binding protein |
| chr18_+_8609402 | 0.04 |
ENST00000329286.6
|
RAB12
|
RAB12, member RAS oncogene family |
| chr22_-_36784035 | 0.04 |
ENST00000216181.5
|
MYH9
|
myosin, heavy chain 9, non-muscle |
| chr1_-_29450399 | 0.04 |
ENST00000521452.1
|
TMEM200B
|
transmembrane protein 200B |
| chr10_-_60027642 | 0.04 |
ENST00000373935.3
|
IPMK
|
inositol polyphosphate multikinase |
| chr14_+_57735614 | 0.04 |
ENST00000261558.3
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr9_-_27573392 | 0.03 |
ENST00000380003.3
|
C9orf72
|
chromosome 9 open reading frame 72 |
| chr1_+_94883931 | 0.03 |
ENST00000394233.2
ENST00000454898.2 ENST00000536817.1 |
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr4_+_52709229 | 0.03 |
ENST00000334635.5
ENST00000381441.3 ENST00000381437.4 |
DCUN1D4
|
DCN1, defective in cullin neddylation 1, domain containing 4 |
| chr10_+_5726764 | 0.03 |
ENST00000328090.5
ENST00000496681.1 |
FAM208B
|
family with sequence similarity 208, member B |
| chr14_+_103851712 | 0.03 |
ENST00000440884.3
ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3
|
MAP/microtubule affinity-regulating kinase 3 |
| chr15_-_52821247 | 0.03 |
ENST00000399231.3
ENST00000399233.2 |
MYO5A
|
myosin VA (heavy chain 12, myoxin) |
| chr10_+_17686124 | 0.03 |
ENST00000377524.3
|
STAM
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr13_-_41240717 | 0.03 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr10_-_32345305 | 0.03 |
ENST00000302418.4
|
KIF5B
|
kinesin family member 5B |
| chr17_-_27278304 | 0.03 |
ENST00000577226.1
|
PHF12
|
PHD finger protein 12 |
| chr18_+_57567180 | 0.03 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr3_+_196295482 | 0.03 |
ENST00000440469.1
ENST00000311630.6 |
FBXO45
|
F-box protein 45 |
| chrX_-_41782249 | 0.03 |
ENST00000442742.2
ENST00000421587.2 ENST00000378166.4 ENST00000318588.9 ENST00000361962.4 |
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr4_+_144257915 | 0.03 |
ENST00000262995.4
|
GAB1
|
GRB2-associated binding protein 1 |
| chrX_-_102942961 | 0.03 |
ENST00000434230.1
ENST00000418819.1 ENST00000360458.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr1_-_184723942 | 0.03 |
ENST00000318130.8
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr12_+_14518598 | 0.03 |
ENST00000261168.4
ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr10_-_27529716 | 0.03 |
ENST00000375897.3
ENST00000396271.3 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr10_+_31608054 | 0.03 |
ENST00000320985.10
ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1
|
zinc finger E-box binding homeobox 1 |
| chr7_+_6414128 | 0.03 |
ENST00000348035.4
ENST00000356142.4 |
RAC1
|
ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
| chr3_-_123603137 | 0.03 |
ENST00000360304.3
ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK
|
myosin light chain kinase |
| chr2_+_198380289 | 0.03 |
ENST00000233892.4
ENST00000409916.1 |
MOB4
|
MOB family member 4, phocein |
| chr15_+_66161871 | 0.03 |
ENST00000569896.1
|
RAB11A
|
RAB11A, member RAS oncogene family |
| chr8_+_61429416 | 0.03 |
ENST00000262646.7
ENST00000531289.1 |
RAB2A
|
RAB2A, member RAS oncogene family |
| chr7_-_151217001 | 0.03 |
ENST00000262187.5
|
RHEB
|
Ras homolog enriched in brain |
| chr10_+_98592009 | 0.03 |
ENST00000540664.1
ENST00000371103.3 |
LCOR
|
ligand dependent nuclear receptor corepressor |
| chr6_-_136610911 | 0.03 |
ENST00000530767.1
ENST00000527759.1 ENST00000527536.1 ENST00000529826.1 ENST00000531224.1 ENST00000353331.4 |
BCLAF1
|
BCL2-associated transcription factor 1 |
| chr9_+_101867359 | 0.03 |
ENST00000374994.4
|
TGFBR1
|
transforming growth factor, beta receptor 1 |
| chr6_+_13615554 | 0.03 |
ENST00000451315.2
|
NOL7
|
nucleolar protein 7, 27kDa |
| chr1_+_218519577 | 0.03 |
ENST00000366930.4
ENST00000366929.4 |
TGFB2
|
transforming growth factor, beta 2 |
| chrX_-_80065146 | 0.03 |
ENST00000373275.4
|
BRWD3
|
bromodomain and WD repeat domain containing 3 |
| chr12_-_101801505 | 0.03 |
ENST00000539055.1
ENST00000551688.1 ENST00000551671.1 ENST00000261636.8 |
ARL1
|
ADP-ribosylation factor-like 1 |
| chr15_+_101142722 | 0.03 |
ENST00000332783.7
ENST00000558747.1 ENST00000343276.4 |
ASB7
|
ankyrin repeat and SOCS box containing 7 |
| chr2_+_187454749 | 0.02 |
ENST00000261023.3
ENST00000374907.3 |
ITGAV
|
integrin, alpha V |
| chr5_-_90679145 | 0.02 |
ENST00000265138.3
|
ARRDC3
|
arrestin domain containing 3 |
| chr18_-_23670546 | 0.02 |
ENST00000542743.1
ENST00000545952.1 ENST00000539849.1 ENST00000415083.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr11_+_34642656 | 0.02 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr1_-_91487013 | 0.02 |
ENST00000347275.5
ENST00000370440.1 |
ZNF644
|
zinc finger protein 644 |
| chr12_+_110719032 | 0.02 |
ENST00000395494.2
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr5_+_112043186 | 0.02 |
ENST00000509732.1
ENST00000457016.1 ENST00000507379.1 |
APC
|
adenomatous polyposis coli |
| chr11_+_13299186 | 0.02 |
ENST00000527998.1
ENST00000396441.3 ENST00000533520.1 ENST00000529825.1 ENST00000389707.4 ENST00000401424.1 ENST00000529388.1 ENST00000530357.1 ENST00000403290.1 ENST00000361003.4 ENST00000389708.3 ENST00000403510.3 ENST00000482049.1 |
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr5_-_156486120 | 0.02 |
ENST00000522693.1
|
HAVCR1
|
hepatitis A virus cellular receptor 1 |
| chrX_+_37208521 | 0.02 |
ENST00000378628.4
|
PRRG1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr11_+_102188272 | 0.02 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr11_-_85780086 | 0.02 |
ENST00000532317.1
ENST00000528256.1 ENST00000526033.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr1_+_93913713 | 0.02 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr2_-_65357225 | 0.02 |
ENST00000398529.3
ENST00000409751.1 ENST00000356214.7 ENST00000409892.1 ENST00000409784.3 |
RAB1A
|
RAB1A, member RAS oncogene family |
| chr6_+_53659746 | 0.02 |
ENST00000370888.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chr14_-_81687197 | 0.02 |
ENST00000553612.1
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chr9_-_80646374 | 0.02 |
ENST00000286548.4
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr5_-_39074479 | 0.02 |
ENST00000514735.1
ENST00000296782.5 ENST00000357387.3 |
RICTOR
|
RPTOR independent companion of MTOR, complex 2 |
| chr2_+_42396472 | 0.02 |
ENST00000318522.5
ENST00000402711.2 |
EML4
|
echinoderm microtubule associated protein like 4 |
| chr1_-_86622421 | 0.02 |
ENST00000370571.2
|
COL24A1
|
collagen, type XXIV, alpha 1 |
| chr5_+_145583156 | 0.02 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr10_-_65225722 | 0.02 |
ENST00000399251.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr14_-_31676964 | 0.02 |
ENST00000553700.1
|
HECTD1
|
HECT domain containing E3 ubiquitin protein ligase 1 |
| chr18_+_67956135 | 0.02 |
ENST00000397942.3
|
SOCS6
|
suppressor of cytokine signaling 6 |
| chr4_+_38869410 | 0.02 |
ENST00000358869.2
|
FAM114A1
|
family with sequence similarity 114, member A1 |
| chr12_+_66217911 | 0.02 |
ENST00000403681.2
|
HMGA2
|
high mobility group AT-hook 2 |
| chr1_-_205719295 | 0.02 |
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr10_-_119806085 | 0.02 |
ENST00000355624.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I) |
| chr1_-_78148324 | 0.02 |
ENST00000370801.3
ENST00000433749.1 |
ZZZ3
|
zinc finger, ZZ-type containing 3 |
| chr4_-_129208940 | 0.02 |
ENST00000296425.5
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr5_-_32444828 | 0.02 |
ENST00000265069.8
|
ZFR
|
zinc finger RNA binding protein |
| chr2_+_135676381 | 0.02 |
ENST00000537343.1
ENST00000295238.6 ENST00000264157.5 |
CCNT2
|
cyclin T2 |
| chr2_+_198365122 | 0.02 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr2_-_86790593 | 0.02 |
ENST00000263856.4
ENST00000409225.2 |
CHMP3
|
charged multivesicular body protein 3 |
| chr9_+_36190853 | 0.02 |
ENST00000433436.2
ENST00000538225.1 ENST00000540080.1 |
CLTA
|
clathrin, light chain A |
| chr11_+_70244510 | 0.02 |
ENST00000346329.3
ENST00000301843.8 ENST00000376561.3 |
CTTN
|
cortactin |
| chr15_+_38544476 | 0.02 |
ENST00000299084.4
|
SPRED1
|
sprouty-related, EVH1 domain containing 1 |
| chr13_+_49550015 | 0.02 |
ENST00000492622.2
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr2_+_9346892 | 0.02 |
ENST00000281419.3
ENST00000315273.4 |
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr11_+_58346584 | 0.02 |
ENST00000316059.6
|
ZFP91
|
ZFP91 zinc finger protein |
| chr19_+_16187085 | 0.02 |
ENST00000300933.4
|
TPM4
|
tropomyosin 4 |
| chr6_+_34857019 | 0.02 |
ENST00000360359.3
ENST00000535627.1 |
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A |
| chr2_-_61765315 | 0.01 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr6_-_134639180 | 0.01 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr1_+_78245303 | 0.01 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chr11_-_76381781 | 0.01 |
ENST00000260061.5
ENST00000404995.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr13_-_31038370 | 0.01 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr12_+_65004292 | 0.01 |
ENST00000542104.1
ENST00000336061.2 |
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr11_+_120207787 | 0.01 |
ENST00000397843.2
ENST00000356641.3 |
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr1_+_7831323 | 0.01 |
ENST00000054666.6
|
VAMP3
|
vesicle-associated membrane protein 3 |
| chr1_+_87170247 | 0.01 |
ENST00000370558.4
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1 |
| chr8_-_38239732 | 0.01 |
ENST00000534155.1
ENST00000433384.2 ENST00000317025.8 ENST00000316985.3 |
WHSC1L1
|
Wolf-Hirschhorn syndrome candidate 1-like 1 |
| chr2_-_68479614 | 0.01 |
ENST00000234310.3
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chr20_-_45984401 | 0.01 |
ENST00000311275.7
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr7_-_105752651 | 0.01 |
ENST00000470347.1
ENST00000455385.2 |
SYPL1
|
synaptophysin-like 1 |
| chr18_-_33647487 | 0.01 |
ENST00000590898.1
ENST00000357384.4 ENST00000319040.6 ENST00000588737.1 ENST00000399022.4 |
RPRD1A
|
regulation of nuclear pre-mRNA domain containing 1A |
| chr3_+_128968437 | 0.01 |
ENST00000314797.6
|
COPG1
|
coatomer protein complex, subunit gamma 1 |
| chr15_+_71389281 | 0.01 |
ENST00000355327.3
|
THSD4
|
thrombospondin, type I, domain containing 4 |
| chr5_-_53606396 | 0.01 |
ENST00000504924.1
ENST00000507646.2 ENST00000502271.1 |
ARL15
|
ADP-ribosylation factor-like 15 |
| chr20_-_61569227 | 0.01 |
ENST00000266070.4
ENST00000395335.2 ENST00000266071.5 |
DIDO1
|
death inducer-obliterator 1 |
| chr17_+_44668035 | 0.01 |
ENST00000398238.4
ENST00000225282.8 |
NSF
|
N-ethylmaleimide-sensitive factor |
| chr5_+_53813536 | 0.01 |
ENST00000343017.6
ENST00000381410.4 ENST00000326277.3 |
SNX18
|
sorting nexin 18 |
| chr15_+_79165151 | 0.01 |
ENST00000331268.5
|
MORF4L1
|
mortality factor 4 like 1 |
| chr5_+_134181625 | 0.01 |
ENST00000394976.3
|
C5orf24
|
chromosome 5 open reading frame 24 |
| chr11_+_126152954 | 0.01 |
ENST00000392679.1
|
TIRAP
|
toll-interleukin 1 receptor (TIR) domain containing adaptor protein |
| chrX_+_44732757 | 0.01 |
ENST00000377967.4
ENST00000536777.1 ENST00000382899.4 ENST00000543216.1 |
KDM6A
|
lysine (K)-specific demethylase 6A |
| chr6_+_163835669 | 0.01 |
ENST00000453779.2
ENST00000275262.7 ENST00000392127.2 ENST00000361752.3 |
QKI
|
QKI, KH domain containing, RNA binding |
| chr17_+_67410832 | 0.01 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr7_-_6523755 | 0.01 |
ENST00000436575.1
ENST00000258739.4 |
DAGLB
KDELR2
|
diacylglycerol lipase, beta KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr7_+_21467642 | 0.01 |
ENST00000222584.3
ENST00000432066.2 |
SP4
|
Sp4 transcription factor |
| chr14_-_35182994 | 0.01 |
ENST00000341223.3
|
CFL2
|
cofilin 2 (muscle) |
| chr10_+_97803151 | 0.01 |
ENST00000403870.3
ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ
|
cyclin J |
| chr2_+_179345173 | 0.01 |
ENST00000234453.5
|
PLEKHA3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr2_-_153032484 | 0.01 |
ENST00000263904.4
|
STAM2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
| chr2_-_39664405 | 0.01 |
ENST00000341681.5
ENST00000263881.3 |
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr1_-_31538517 | 0.01 |
ENST00000440538.2
ENST00000423018.2 ENST00000424085.2 ENST00000426105.2 ENST00000257075.5 ENST00000373747.3 ENST00000525843.1 ENST00000373742.2 |
PUM1
|
pumilio RNA-binding family member 1 |
| chr1_+_224544552 | 0.01 |
ENST00000465271.1
ENST00000366858.3 |
CNIH4
|
cornichon family AMPA receptor auxiliary protein 4 |
| chr6_-_132834184 | 0.01 |
ENST00000367941.2
ENST00000367937.4 |
STX7
|
syntaxin 7 |
| chr7_-_123389104 | 0.01 |
ENST00000223023.4
|
WASL
|
Wiskott-Aldrich syndrome-like |
| chr2_+_204192942 | 0.01 |
ENST00000295851.5
ENST00000261017.5 |
ABI2
|
abl-interactor 2 |
| chr1_-_155532484 | 0.01 |
ENST00000368346.3
ENST00000548830.1 |
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr5_-_142783175 | 0.01 |
ENST00000231509.3
ENST00000394464.2 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr5_-_173043591 | 0.01 |
ENST00000285908.5
ENST00000480951.1 ENST00000311086.4 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr4_+_87856129 | 0.01 |
ENST00000395146.4
ENST00000507468.1 |
AFF1
|
AF4/FMR2 family, member 1 |
| chr5_-_114961858 | 0.01 |
ENST00000282382.4
ENST00000456936.3 ENST00000408996.4 |
TMED7-TICAM2
TMED7
TICAM2
|
TMED7-TICAM2 readthrough transmembrane emp24 protein transport domain containing 7 toll-like receptor adaptor molecule 2 |
| chr10_+_101292684 | 0.01 |
ENST00000344586.7
|
NKX2-3
|
NK2 homeobox 3 |
| chr1_-_226924980 | 0.01 |
ENST00000272117.3
|
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr4_+_699537 | 0.01 |
ENST00000419774.1
ENST00000362003.5 ENST00000400151.2 ENST00000427463.1 ENST00000470161.2 |
PCGF3
|
polycomb group ring finger 3 |
| chr8_+_9413410 | 0.01 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr18_+_43753974 | 0.01 |
ENST00000282059.6
ENST00000321319.6 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr7_-_44924939 | 0.00 |
ENST00000395699.2
|
PURB
|
purine-rich element binding protein B |
| chr1_+_114472222 | 0.00 |
ENST00000369558.1
ENST00000369561.4 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr6_-_30585009 | 0.00 |
ENST00000376511.2
|
PPP1R10
|
protein phosphatase 1, regulatory subunit 10 |
| chr2_+_210444142 | 0.00 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr14_-_93799360 | 0.00 |
ENST00000334746.5
ENST00000554565.1 ENST00000298896.3 |
BTBD7
|
BTB (POZ) domain containing 7 |
| chr12_+_8850471 | 0.00 |
ENST00000535829.1
ENST00000357529.3 |
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr1_+_66797687 | 0.00 |
ENST00000371045.5
ENST00000531025.1 ENST00000526197.1 |
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr9_+_116638562 | 0.00 |
ENST00000374126.5
ENST00000288466.7 |
ZNF618
|
zinc finger protein 618 |
| chr6_-_30712313 | 0.00 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr17_-_62340581 | 0.00 |
ENST00000258991.3
ENST00000583738.1 ENST00000584379.1 |
TEX2
|
testis expressed 2 |
| chr16_-_74700737 | 0.00 |
ENST00000576652.1
ENST00000572337.1 ENST00000571750.1 ENST00000572990.1 ENST00000361070.4 |
RFWD3
|
ring finger and WD repeat domain 3 |
| chr6_-_138893661 | 0.00 |
ENST00000427025.2
|
NHSL1
|
NHS-like 1 |
| chr11_+_122709200 | 0.00 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr13_+_28813645 | 0.00 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr17_+_2699697 | 0.00 |
ENST00000254695.8
ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2
|
RAP1 GTPase activating protein 2 |
| chr2_-_161350305 | 0.00 |
ENST00000348849.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr14_+_65381079 | 0.00 |
ENST00000549115.1
ENST00000607599.1 ENST00000548752.2 ENST00000359118.2 ENST00000552002.2 ENST00000551947.1 ENST00000551093.1 ENST00000542227.1 ENST00000447296.2 ENST00000549987.1 |
CHURC1
FNTB
CHURC1-FNTB
|
churchill domain containing 1 farnesyltransferase, CAAX box, beta CHURC1-FNTB readthrough |
| chr12_-_117628333 | 0.00 |
ENST00000427718.2
|
FBXO21
|
F-box protein 21 |
| chr3_+_93781728 | 0.00 |
ENST00000314622.4
|
NSUN3
|
NOP2/Sun domain family, member 3 |
| chr5_+_95066823 | 0.00 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr17_-_48474828 | 0.00 |
ENST00000576448.1
ENST00000225972.7 |
LRRC59
|
leucine rich repeat containing 59 |
| chr11_+_118307179 | 0.00 |
ENST00000534358.1
ENST00000531904.2 ENST00000389506.5 ENST00000354520.4 |
KMT2A
|
lysine (K)-specific methyltransferase 2A |
| chr14_-_21737610 | 0.00 |
ENST00000320084.7
ENST00000449098.1 ENST00000336053.6 |
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr20_+_8112824 | 0.00 |
ENST00000378641.3
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr3_+_100428268 | 0.00 |
ENST00000240851.4
|
TFG
|
TRK-fused gene |
| chrX_+_147582130 | 0.00 |
ENST00000370460.2
ENST00000370457.5 |
AFF2
|
AF4/FMR2 family, member 2 |
| chr9_-_115095883 | 0.00 |
ENST00000450374.1
ENST00000374255.2 ENST00000334318.6 ENST00000374257.1 |
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr7_+_20370746 | 0.00 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr6_-_74363803 | 0.00 |
ENST00000355773.5
|
SLC17A5
|
solute carrier family 17 (acidic sugar transporter), member 5 |
| chr11_+_57520715 | 0.00 |
ENST00000524630.1
ENST00000529919.1 ENST00000399039.4 ENST00000533189.1 |
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr6_+_7107999 | 0.00 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr1_+_213031570 | 0.00 |
ENST00000366971.4
|
FLVCR1
|
feline leukemia virus subgroup C cellular receptor 1 |
| chr9_-_127703333 | 0.00 |
ENST00000373555.4
|
GOLGA1
|
golgin A1 |
| chr5_-_133512683 | 0.00 |
ENST00000353411.6
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr4_-_16077741 | 0.00 |
ENST00000447510.2
ENST00000540805.1 ENST00000539194.1 |
PROM1
|
prominin 1 |
| chr20_-_3996036 | 0.00 |
ENST00000336095.6
|
RNF24
|
ring finger protein 24 |
| chr1_-_179198702 | 0.00 |
ENST00000502732.1
|
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
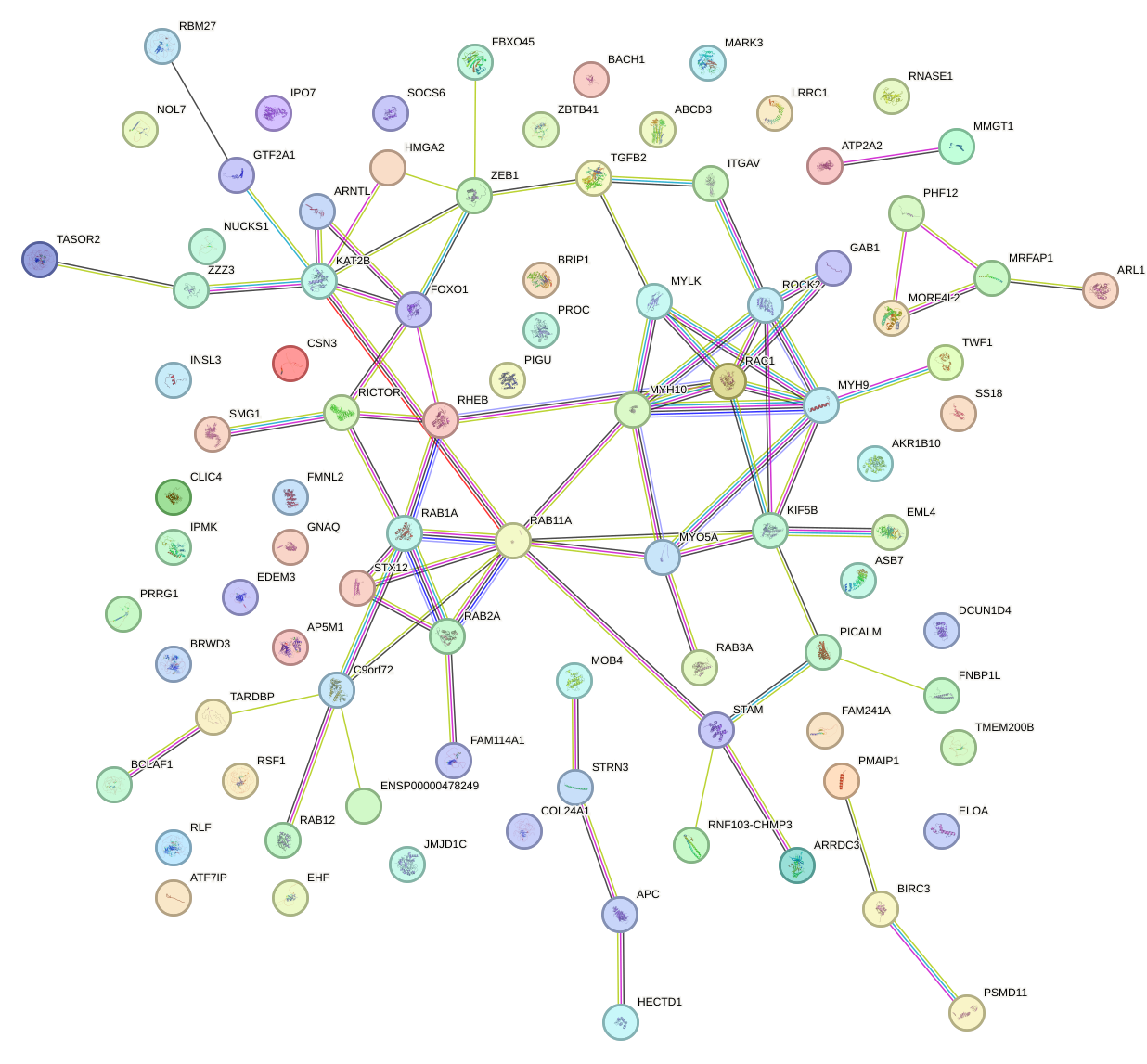
Network of associatons between targets according to the STRING database.
First level regulatory network of GUAGUGU
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.0 | GO:0031213 | RSF complex(GO:0031213) |