Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
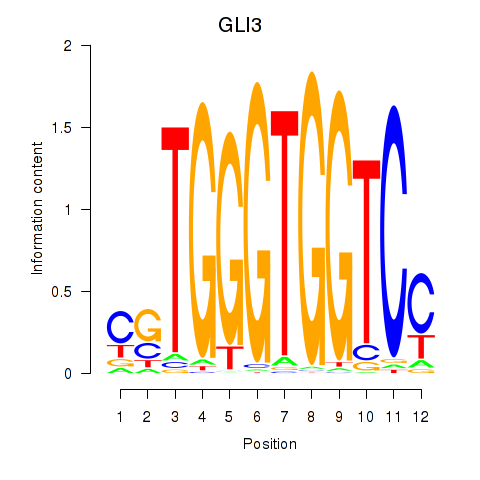
Results for GLI3
Z-value: 1.86
Transcription factors associated with GLI3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLI3
|
ENSG00000106571.8 | GLI family zinc finger 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLI3 | hg19_v2_chr7_-_42276612_42276782 | -0.75 | 2.5e-01 | Click! |
Activity profile of GLI3 motif
Sorted Z-values of GLI3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_50148087 | 1.59 |
ENST00000601038.1
ENST00000595242.1 |
SCAF1
|
SR-related CTD-associated factor 1 |
| chr19_-_46145696 | 1.22 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr19_+_51152702 | 1.20 |
ENST00000425202.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr16_+_29832634 | 0.97 |
ENST00000565164.1
ENST00000570234.1 |
MVP
|
major vault protein |
| chr19_-_30199516 | 0.97 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr19_-_50381606 | 0.93 |
ENST00000391830.1
|
AKT1S1
|
AKT1 substrate 1 (proline-rich) |
| chr7_+_76101379 | 0.92 |
ENST00000429179.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr11_+_76493294 | 0.92 |
ENST00000533752.1
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr16_+_70695570 | 0.86 |
ENST00000597002.1
|
FLJ00418
|
FLJ00418 |
| chr20_+_62327996 | 0.85 |
ENST00000369996.1
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy |
| chr19_+_782755 | 0.83 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr19_+_676385 | 0.83 |
ENST00000166139.4
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein) |
| chr7_-_994302 | 0.82 |
ENST00000265846.5
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr19_+_18283959 | 0.81 |
ENST00000597802.2
|
IFI30
|
interferon, gamma-inducible protein 30 |
| chr14_+_105559784 | 0.81 |
ENST00000548104.1
|
RP11-44N21.1
|
RP11-44N21.1 |
| chr11_+_64053311 | 0.77 |
ENST00000540370.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr11_+_64073699 | 0.77 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr16_+_30675654 | 0.74 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr12_-_57030096 | 0.74 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr6_-_33756867 | 0.74 |
ENST00000293760.5
|
LEMD2
|
LEM domain containing 2 |
| chr14_+_94577074 | 0.74 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr17_-_7307358 | 0.72 |
ENST00000576017.1
ENST00000302422.3 ENST00000535512.1 |
TMEM256
TMEM256-PLSCR3
|
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr11_+_393428 | 0.72 |
ENST00000533249.1
ENST00000527442.1 |
PKP3
|
plakophilin 3 |
| chr8_+_94752349 | 0.72 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr19_-_10464570 | 0.71 |
ENST00000529739.1
|
TYK2
|
tyrosine kinase 2 |
| chr5_+_149340339 | 0.67 |
ENST00000433184.1
|
SLC26A2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr8_-_144691718 | 0.66 |
ENST00000377579.3
ENST00000433751.1 ENST00000220966.6 |
PYCRL
|
pyrroline-5-carboxylate reductase-like |
| chr1_-_1310530 | 0.65 |
ENST00000338370.3
ENST00000321751.5 ENST00000378853.3 |
AURKAIP1
|
aurora kinase A interacting protein 1 |
| chr3_+_50388126 | 0.65 |
ENST00000425346.1
ENST00000424512.1 ENST00000232508.5 ENST00000418577.1 ENST00000606589.1 |
CYB561D2
XXcos-LUCA11.5
|
cytochrome b561 family, member D2 Uncharacterized protein |
| chr11_-_111649074 | 0.64 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr1_-_1310870 | 0.64 |
ENST00000338338.5
|
AURKAIP1
|
aurora kinase A interacting protein 1 |
| chr11_-_796197 | 0.63 |
ENST00000530360.1
ENST00000528606.1 ENST00000320230.5 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr19_+_39989535 | 0.63 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr10_+_71389983 | 0.61 |
ENST00000373279.4
|
C10orf35
|
chromosome 10 open reading frame 35 |
| chrX_-_48693955 | 0.61 |
ENST00000218230.5
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr19_-_55895966 | 0.60 |
ENST00000444469.3
|
TMEM238
|
transmembrane protein 238 |
| chr11_-_118789613 | 0.59 |
ENST00000532899.1
|
BCL9L
|
B-cell CLL/lymphoma 9-like |
| chr6_-_27841289 | 0.59 |
ENST00000355981.2
|
HIST1H4L
|
histone cluster 1, H4l |
| chr7_+_99954224 | 0.59 |
ENST00000608825.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
| chr17_+_73089382 | 0.58 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr8_+_22428457 | 0.57 |
ENST00000517962.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr16_-_850723 | 0.57 |
ENST00000248150.4
|
GNG13
|
guanine nucleotide binding protein (G protein), gamma 13 |
| chr17_+_43238438 | 0.56 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr16_-_11681023 | 0.56 |
ENST00000570904.1
ENST00000574701.1 |
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr19_-_6379069 | 0.55 |
ENST00000597721.1
|
PSPN
|
persephin |
| chr11_+_1295809 | 0.55 |
ENST00000598274.1
|
AC136297.1
|
Uncharacterized protein |
| chr19_-_2041159 | 0.54 |
ENST00000589441.1
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr16_+_77233294 | 0.54 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr19_-_13947099 | 0.54 |
ENST00000587762.1
|
MIR24-2
|
microRNA 24-2 |
| chr7_-_2595337 | 0.53 |
ENST00000340611.4
|
BRAT1
|
BRCA1-associated ATM activator 1 |
| chrX_+_153626571 | 0.53 |
ENST00000424325.2
|
RPL10
|
ribosomal protein L10 |
| chr12_-_54582655 | 0.53 |
ENST00000504338.1
ENST00000514685.1 ENST00000504797.1 ENST00000513838.1 ENST00000505128.1 ENST00000337581.3 ENST00000503306.1 ENST00000243112.5 ENST00000514196.1 ENST00000506169.1 ENST00000507904.1 ENST00000508394.2 |
SMUG1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| chr5_-_180237082 | 0.53 |
ENST00000506889.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr19_+_48281842 | 0.53 |
ENST00000509570.2
|
SEPW1
|
selenoprotein W, 1 |
| chr16_+_2820912 | 0.52 |
ENST00000570539.1
|
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr14_+_105155925 | 0.52 |
ENST00000330634.7
ENST00000398337.4 ENST00000392634.4 |
INF2
|
inverted formin, FH2 and WH2 domain containing |
| chr20_+_3190006 | 0.52 |
ENST00000380113.3
ENST00000455664.2 ENST00000399838.3 |
ITPA
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
| chr5_-_137090028 | 0.51 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr14_+_24099318 | 0.51 |
ENST00000432832.2
|
DHRS2
|
dehydrogenase/reductase (SDR family) member 2 |
| chr19_+_45417812 | 0.51 |
ENST00000592535.1
|
APOC1
|
apolipoprotein C-I |
| chr16_+_765092 | 0.51 |
ENST00000568223.2
|
METRN
|
meteorin, glial cell differentiation regulator |
| chr1_-_21978312 | 0.50 |
ENST00000359708.4
ENST00000290101.4 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr5_+_23951673 | 0.50 |
ENST00000507936.1
|
C5orf17
|
chromosome 5 open reading frame 17 |
| chr19_-_33793430 | 0.50 |
ENST00000498907.2
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr14_+_105147464 | 0.50 |
ENST00000540171.2
|
RP11-982M15.6
|
RP11-982M15.6 |
| chr7_+_2685164 | 0.50 |
ENST00000400376.2
|
TTYH3
|
tweety family member 3 |
| chr16_-_4817129 | 0.49 |
ENST00000545009.1
ENST00000219478.6 |
ZNF500
|
zinc finger protein 500 |
| chr22_-_50970919 | 0.49 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr16_+_28875268 | 0.48 |
ENST00000395532.4
|
SH2B1
|
SH2B adaptor protein 1 |
| chr19_+_49713991 | 0.48 |
ENST00000597316.1
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr16_+_70488480 | 0.48 |
ENST00000572784.1
ENST00000574784.1 ENST00000571514.1 ENST00000378912.2 ENST00000428974.2 ENST00000573352.1 ENST00000576453.1 |
FUK
|
fucokinase |
| chr17_+_80193644 | 0.48 |
ENST00000582946.1
|
SLC16A3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr16_+_84209738 | 0.47 |
ENST00000564928.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr5_-_180237445 | 0.47 |
ENST00000393340.3
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr19_+_44037334 | 0.47 |
ENST00000314228.5
|
ZNF575
|
zinc finger protein 575 |
| chr12_+_6977323 | 0.46 |
ENST00000462761.1
|
TPI1
|
triosephosphate isomerase 1 |
| chr20_-_62258394 | 0.46 |
ENST00000370077.1
|
GMEB2
|
glucocorticoid modulatory element binding protein 2 |
| chr12_-_111806892 | 0.46 |
ENST00000547710.1
ENST00000549321.1 ENST00000361483.3 ENST00000392658.5 |
FAM109A
|
family with sequence similarity 109, member A |
| chr19_+_1905365 | 0.46 |
ENST00000329478.2
ENST00000602400.1 ENST00000409472.1 |
ADAT3
SCAMP4
|
adenosine deaminase, tRNA-specific 3 secretory carrier membrane protein 4 |
| chr11_-_568369 | 0.46 |
ENST00000534540.1
ENST00000528245.1 ENST00000500447.1 ENST00000533920.1 |
MIR210HG
|
MIR210 host gene (non-protein coding) |
| chr19_-_4454081 | 0.45 |
ENST00000591919.1
|
UBXN6
|
UBX domain protein 6 |
| chr19_+_39989580 | 0.45 |
ENST00000596614.1
ENST00000205143.4 |
DLL3
|
delta-like 3 (Drosophila) |
| chr7_-_100034060 | 0.45 |
ENST00000292330.2
|
PPP1R35
|
protein phosphatase 1, regulatory subunit 35 |
| chr19_+_17337027 | 0.45 |
ENST00000601529.1
ENST00000600232.1 |
OCEL1
|
occludin/ELL domain containing 1 |
| chr8_-_144890847 | 0.45 |
ENST00000531942.1
|
SCRIB
|
scribbled planar cell polarity protein |
| chr19_+_18699535 | 0.45 |
ENST00000358607.6
|
C19orf60
|
chromosome 19 open reading frame 60 |
| chr9_+_139247484 | 0.45 |
ENST00000429455.1
|
GPSM1
|
G-protein signaling modulator 1 |
| chr16_-_88717482 | 0.44 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr19_-_39832563 | 0.44 |
ENST00000599274.1
|
CTC-246B18.10
|
CTC-246B18.10 |
| chr22_-_21213676 | 0.44 |
ENST00000449120.1
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha |
| chr2_-_74726710 | 0.44 |
ENST00000377566.4
|
LBX2
|
ladybird homeobox 2 |
| chr6_-_85473073 | 0.44 |
ENST00000606621.1
|
TBX18
|
T-box 18 |
| chr19_+_3708338 | 0.44 |
ENST00000590545.1
|
TJP3
|
tight junction protein 3 |
| chr17_+_46184911 | 0.44 |
ENST00000580219.1
ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11
|
sorting nexin 11 |
| chr7_+_33168856 | 0.44 |
ENST00000432983.1
|
BBS9
|
Bardet-Biedl syndrome 9 |
| chr11_-_1785139 | 0.44 |
ENST00000236671.2
|
CTSD
|
cathepsin D |
| chr16_+_58033450 | 0.44 |
ENST00000561743.1
|
USB1
|
U6 snRNA biogenesis 1 |
| chr1_+_180165672 | 0.43 |
ENST00000443059.1
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr5_+_140044261 | 0.43 |
ENST00000358337.5
|
WDR55
|
WD repeat domain 55 |
| chr17_+_73642315 | 0.43 |
ENST00000556126.2
|
SMIM6
|
small integral membrane protein 6 |
| chr16_+_68279207 | 0.43 |
ENST00000413021.2
ENST00000565744.1 ENST00000219345.5 |
PLA2G15
|
phospholipase A2, group XV |
| chr19_+_45165545 | 0.43 |
ENST00000592472.1
ENST00000587729.1 ENST00000585657.1 ENST00000592789.1 ENST00000591979.1 |
CEACAM19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr6_-_26027480 | 0.43 |
ENST00000377364.3
|
HIST1H4B
|
histone cluster 1, H4b |
| chr6_-_33548006 | 0.43 |
ENST00000374467.3
|
BAK1
|
BCL2-antagonist/killer 1 |
| chr19_+_58962971 | 0.43 |
ENST00000336614.4
ENST00000545523.1 ENST00000599194.1 ENST00000598244.1 ENST00000599193.1 ENST00000594214.1 ENST00000391696.1 |
ZNF324B
|
zinc finger protein 324B |
| chr15_+_89164560 | 0.43 |
ENST00000379231.3
ENST00000559528.1 |
AEN
|
apoptosis enhancing nuclease |
| chr8_+_143761874 | 0.43 |
ENST00000301258.4
ENST00000513264.1 |
PSCA
|
prostate stem cell antigen |
| chr17_-_7137857 | 0.42 |
ENST00000005340.5
|
DVL2
|
dishevelled segment polarity protein 2 |
| chr2_+_201981663 | 0.42 |
ENST00000433445.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr6_-_31671058 | 0.42 |
ENST00000538874.1
ENST00000395952.3 |
ABHD16A
|
abhydrolase domain containing 16A |
| chr20_+_43343476 | 0.42 |
ENST00000372868.2
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr8_+_22857048 | 0.42 |
ENST00000251822.6
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr1_+_44440575 | 0.42 |
ENST00000532642.1
ENST00000236067.4 ENST00000471859.2 |
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b |
| chr17_+_76422409 | 0.42 |
ENST00000600087.1
|
AC061992.1
|
Uncharacterized protein |
| chr16_+_88493879 | 0.42 |
ENST00000565624.1
ENST00000437464.1 |
ZNF469
|
zinc finger protein 469 |
| chr3_-_135915146 | 0.42 |
ENST00000473093.1
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr19_-_47922750 | 0.42 |
ENST00000331559.5
|
MEIS3
|
Meis homeobox 3 |
| chr17_+_43239191 | 0.41 |
ENST00000589230.1
|
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr1_+_24117627 | 0.41 |
ENST00000400061.1
|
LYPLA2
|
lysophospholipase II |
| chr17_-_19062424 | 0.40 |
ENST00000399083.1
|
AC007952.6
|
Uncharacterized protein |
| chr8_-_94752946 | 0.40 |
ENST00000519109.1
|
RBM12B
|
RNA binding motif protein 12B |
| chr19_-_54693146 | 0.40 |
ENST00000414665.1
ENST00000453320.1 |
MBOAT7
|
membrane bound O-acyltransferase domain containing 7 |
| chr19_+_4969116 | 0.40 |
ENST00000588337.1
ENST00000159111.4 ENST00000381759.4 |
KDM4B
|
lysine (K)-specific demethylase 4B |
| chr2_-_201936302 | 0.40 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr16_+_68279256 | 0.39 |
ENST00000564827.2
ENST00000566188.1 ENST00000444212.2 ENST00000568082.1 |
PLA2G15
|
phospholipase A2, group XV |
| chr12_+_51632638 | 0.39 |
ENST00000549732.2
|
DAZAP2
|
DAZ associated protein 2 |
| chr14_+_71165292 | 0.39 |
ENST00000553682.1
|
RP6-65G23.1
|
RP6-65G23.1 |
| chr2_-_11272234 | 0.39 |
ENST00000590207.1
ENST00000417697.2 ENST00000396164.1 ENST00000536743.1 ENST00000544306.1 |
AC062028.1
|
AC062028.1 |
| chr10_-_103454876 | 0.39 |
ENST00000331272.7
|
FBXW4
|
F-box and WD repeat domain containing 4 |
| chr19_+_5914213 | 0.38 |
ENST00000222125.5
ENST00000452990.2 ENST00000588865.1 |
CAPS
|
calcyphosine |
| chr16_+_89979826 | 0.38 |
ENST00000555427.1
|
MC1R
|
melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor) |
| chr19_+_37960466 | 0.38 |
ENST00000589725.1
|
ZNF570
|
zinc finger protein 570 |
| chr3_-_125802765 | 0.38 |
ENST00000514891.1
ENST00000512470.1 ENST00000504035.1 ENST00000360370.4 ENST00000513723.1 ENST00000510651.1 ENST00000514333.1 |
SLC41A3
|
solute carrier family 41, member 3 |
| chr11_-_65374430 | 0.38 |
ENST00000532507.1
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr16_-_2260834 | 0.38 |
ENST00000562360.1
ENST00000566018.1 |
BRICD5
|
BRICHOS domain containing 5 |
| chr17_+_21729899 | 0.38 |
ENST00000583708.1
|
UBBP4
|
ubiquitin B pseudogene 4 |
| chr3_-_10052849 | 0.38 |
ENST00000437616.1
ENST00000429065.2 |
AC022007.5
|
AC022007.5 |
| chr17_+_73642486 | 0.37 |
ENST00000579469.1
|
SMIM6
|
small integral membrane protein 6 |
| chr11_+_844067 | 0.37 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr20_-_1165117 | 0.37 |
ENST00000381894.3
|
TMEM74B
|
transmembrane protein 74B |
| chr20_+_814377 | 0.37 |
ENST00000304189.2
ENST00000381939.1 |
FAM110A
|
family with sequence similarity 110, member A |
| chr22_+_24990746 | 0.37 |
ENST00000456869.1
ENST00000411974.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr19_+_17337007 | 0.37 |
ENST00000215061.4
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr5_-_180236811 | 0.37 |
ENST00000446023.2
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr11_+_123301012 | 0.37 |
ENST00000533341.1
|
AP000783.1
|
Uncharacterized protein |
| chr22_+_20905422 | 0.37 |
ENST00000424287.1
ENST00000423862.1 |
MED15
|
mediator complex subunit 15 |
| chr8_-_145980808 | 0.37 |
ENST00000525191.1
|
ZNF251
|
zinc finger protein 251 |
| chr17_+_17380294 | 0.36 |
ENST00000268711.3
ENST00000580462.1 |
MED9
|
mediator complex subunit 9 |
| chr14_-_21566731 | 0.36 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr8_-_131028782 | 0.35 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr5_+_150157444 | 0.35 |
ENST00000526627.1
|
SMIM3
|
small integral membrane protein 3 |
| chr22_-_42765174 | 0.35 |
ENST00000432473.1
ENST00000412060.1 ENST00000424852.1 |
Z83851.1
|
Z83851.1 |
| chr2_-_74648702 | 0.35 |
ENST00000518863.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr17_-_79633590 | 0.35 |
ENST00000374741.3
ENST00000571503.1 |
OXLD1
|
oxidoreductase-like domain containing 1 |
| chr17_+_41158742 | 0.35 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr10_-_112255945 | 0.35 |
ENST00000609514.1
ENST00000607952.1 |
RP11-525A16.4
|
RP11-525A16.4 |
| chr19_+_7599128 | 0.35 |
ENST00000545201.2
|
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr12_+_112191694 | 0.35 |
ENST00000546840.2
|
RP11-162P23.2
|
RP11-162P23.2 |
| chr19_-_42931567 | 0.35 |
ENST00000244289.4
|
LIPE
|
lipase, hormone-sensitive |
| chr15_+_29211570 | 0.35 |
ENST00000558804.1
|
APBA2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr8_+_21916710 | 0.35 |
ENST00000523266.1
ENST00000519907.1 |
DMTN
|
dematin actin binding protein |
| chr3_-_128840604 | 0.35 |
ENST00000476465.1
ENST00000315150.5 ENST00000393304.1 ENST00000393308.1 ENST00000393307.1 ENST00000393305.1 |
RAB43
|
RAB43, member RAS oncogene family |
| chr1_+_11724167 | 0.35 |
ENST00000376753.4
|
FBXO6
|
F-box protein 6 |
| chr5_-_180242534 | 0.35 |
ENST00000333055.3
ENST00000513431.1 |
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr17_-_202579 | 0.34 |
ENST00000577079.1
ENST00000331302.7 ENST00000536489.2 |
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr11_-_69294647 | 0.34 |
ENST00000542064.1
|
AP000439.3
|
AP000439.3 |
| chr1_+_17248418 | 0.34 |
ENST00000375541.5
|
CROCC
|
ciliary rootlet coiled-coil, rootletin |
| chr17_+_7210898 | 0.34 |
ENST00000572815.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr11_+_832944 | 0.34 |
ENST00000322008.4
ENST00000397421.1 ENST00000529810.1 ENST00000526693.1 ENST00000525333.1 ENST00000524748.1 ENST00000527341.1 |
CD151
|
CD151 molecule (Raph blood group) |
| chr16_+_2880254 | 0.34 |
ENST00000570670.1
|
ZG16B
|
zymogen granule protein 16B |
| chr8_+_21916680 | 0.34 |
ENST00000358242.3
ENST00000415253.1 |
DMTN
|
dematin actin binding protein |
| chr17_-_7761256 | 0.34 |
ENST00000575208.1
|
LSMD1
|
LSM domain containing 1 |
| chr6_-_33547975 | 0.34 |
ENST00000442998.2
ENST00000360661.5 |
BAK1
|
BCL2-antagonist/killer 1 |
| chr4_-_40632757 | 0.34 |
ENST00000511902.1
ENST00000505220.1 |
RBM47
|
RNA binding motif protein 47 |
| chr19_-_54693401 | 0.34 |
ENST00000338624.6
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7 |
| chr4_+_189376725 | 0.34 |
ENST00000510832.1
ENST00000510005.1 ENST00000503458.1 |
LINC01060
|
long intergenic non-protein coding RNA 1060 |
| chr8_-_145331153 | 0.34 |
ENST00000377412.4
|
KM-PA-2
|
KM-PA-2 protein; Uncharacterized protein |
| chr22_-_38349552 | 0.34 |
ENST00000422191.1
ENST00000249079.2 ENST00000418863.1 ENST00000403305.1 ENST00000403026.1 |
C22orf23
|
chromosome 22 open reading frame 23 |
| chr19_+_7599597 | 0.34 |
ENST00000414982.3
ENST00000450331.3 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr19_-_54618650 | 0.33 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr16_-_68057770 | 0.33 |
ENST00000332395.5
|
DDX28
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 |
| chr22_-_50970506 | 0.33 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr11_-_111649015 | 0.33 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr2_+_201981527 | 0.33 |
ENST00000441224.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr7_-_56119156 | 0.33 |
ENST00000421312.1
ENST00000416592.1 |
PSPH
|
phosphoserine phosphatase |
| chr17_+_46018872 | 0.33 |
ENST00000583599.1
ENST00000434554.2 ENST00000225573.4 ENST00000544840.1 ENST00000534893.1 |
PNPO
|
pyridoxamine 5'-phosphate oxidase |
| chrX_+_23685653 | 0.33 |
ENST00000379331.3
|
PRDX4
|
peroxiredoxin 4 |
| chr11_+_69061594 | 0.33 |
ENST00000441339.2
ENST00000308946.3 ENST00000535407.1 |
MYEOV
|
myeloma overexpressed |
| chr20_-_22566089 | 0.33 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr2_+_242498135 | 0.33 |
ENST00000318407.3
|
BOK
|
BCL2-related ovarian killer |
| chr6_-_30685214 | 0.33 |
ENST00000425072.1
|
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr1_+_63788730 | 0.33 |
ENST00000371116.2
|
FOXD3
|
forkhead box D3 |
| chr20_+_5986756 | 0.32 |
ENST00000452938.1
|
CRLS1
|
cardiolipin synthase 1 |
| chr15_+_89164520 | 0.32 |
ENST00000332810.3
|
AEN
|
apoptosis enhancing nuclease |
| chr15_+_45694567 | 0.32 |
ENST00000559860.1
|
SPATA5L1
|
spermatogenesis associated 5-like 1 |
| chr21_-_45079341 | 0.32 |
ENST00000443485.1
ENST00000291560.2 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chrX_+_152240819 | 0.32 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chr17_-_655278 | 0.32 |
ENST00000574958.1
ENST00000437269.1 ENST00000573482.1 |
GEMIN4
|
gem (nuclear organelle) associated protein 4 |
| chr11_+_71939512 | 0.32 |
ENST00000540329.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr19_+_39897453 | 0.32 |
ENST00000597629.1
ENST00000248673.3 ENST00000594045.1 ENST00000594442.1 |
ZFP36
|
ZFP36 ring finger protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of GLI3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.3 | 0.8 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.2 | 2.0 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.2 | 0.8 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.2 | 0.6 | GO:0050823 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.2 | 0.7 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 0.5 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.2 | 0.5 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.2 | 0.7 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.2 | 0.6 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.2 | 0.6 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 0.6 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 0.3 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.1 | 0.4 | GO:0060829 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.7 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 1.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.5 | GO:1990736 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.1 | 0.3 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.3 | GO:0071810 | circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.1 | 0.5 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.1 | 0.4 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.4 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.3 | GO:1904976 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.1 | 0.7 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.5 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 0.1 | 0.3 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.4 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) |
| 0.1 | 0.4 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.4 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.4 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 0.3 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.1 | 0.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.9 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.3 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.6 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 1.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 0.4 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 1.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.5 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.1 | 0.7 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.1 | 0.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.7 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.3 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.1 | 0.2 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.1 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.5 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.7 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.2 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.1 | 0.3 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.1 | 0.3 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.3 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.7 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.3 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.3 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.1 | 0.8 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.2 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.2 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:0002880 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) positive regulation of chronic inflammatory response(GO:0002678) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.1 | 0.6 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.2 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.2 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.6 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.2 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.2 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.4 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.3 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 | 0.3 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.1 | 0.3 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.5 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.3 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.4 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.5 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 1.8 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 1.6 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 0.1 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.3 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.2 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 1.0 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.1 | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.2 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.3 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0019322 | pentose-phosphate shunt, oxidative branch(GO:0009051) pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.1 | GO:2001301 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:1904729 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of Cdc42 protein signal transduction(GO:0032489) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.8 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.2 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.0 | 0.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0097473 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.5 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.5 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.9 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.7 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.2 | GO:1901526 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.0 | GO:0042634 | regulation of hair cycle(GO:0042634) |
| 0.0 | 0.7 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.4 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.2 | GO:0070981 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.0 | 0.1 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.5 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.7 | GO:0034033 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.1 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.2 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.2 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.5 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.2 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.3 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.5 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.2 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.5 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.2 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.3 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.1 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.1 | GO:0042063 | glial cell migration(GO:0008347) gliogenesis(GO:0042063) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.5 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.9 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.2 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.0 | 0.2 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.2 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:1902568 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.0 | GO:1902824 | regulation of late endosome to lysosome transport(GO:1902822) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.2 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.3 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.0 | GO:0043385 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.0 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.6 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.2 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 1.1 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.0 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.3 | GO:0015684 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.4 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.3 | GO:0046174 | polyol catabolic process(GO:0046174) |
| 0.0 | 0.4 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.4 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.2 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.0 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.2 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.0 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0090402 | senescence-associated heterochromatin focus assembly(GO:0035986) oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.0 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:0010989 | regulation of low-density lipoprotein particle clearance(GO:0010988) negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.0 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.0 | 0.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.2 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.0 | 0.5 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.0 | 0.5 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) positive regulation of mitochondrial translation(GO:0070131) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.1 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.3 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 0.4 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.1 | 0.6 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.6 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 1.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.3 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.7 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.9 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.4 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.2 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.0 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.8 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.4 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.7 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.4 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.3 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0030681 | multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.8 | GO:0044306 | neuron projection terminus(GO:0044306) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 0.6 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.1 | 0.4 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.1 | 0.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.4 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.4 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.8 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.6 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.1 | 0.5 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.5 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.3 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.1 | 0.5 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.4 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 1.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.3 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.1 | 0.3 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 0.3 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.1 | 0.3 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.1 | 0.5 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.3 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.4 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 0.9 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.1 | 0.8 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.3 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.3 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 0.1 | GO:0098847 | sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 0.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.7 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 1.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.2 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 0.2 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.2 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 1.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.6 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 0.2 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.1 | 2.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.2 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.4 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.2 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.0 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.9 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0047977 | linoleate 13S-lipoxygenase activity(GO:0016165) hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.1 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 1.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.0 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.2 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.2 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.6 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) type II activin receptor binding(GO:0070699) |
| 0.0 | 1.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.3 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.1 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.0 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 1.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.0 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0015193 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.4 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 2.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.1 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.8 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |