Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
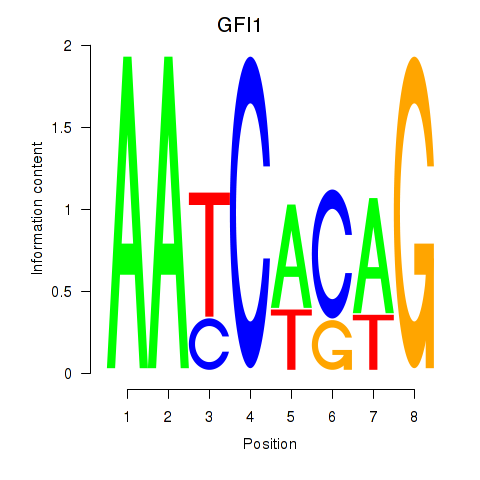
Results for GFI1
Z-value: 0.67
Transcription factors associated with GFI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GFI1
|
ENSG00000162676.7 | growth factor independent 1 transcriptional repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GFI1 | hg19_v2_chr1_-_92951607_92951661 | 0.11 | 8.9e-01 | Click! |
Activity profile of GFI1 motif
Sorted Z-values of GFI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_36602104 | 0.28 |
ENST00000585332.1
ENST00000262637.4 |
OVOL3
|
ovo-like zinc finger 3 |
| chr11_-_4629367 | 0.28 |
ENST00000533021.1
|
TRIM68
|
tripartite motif containing 68 |
| chr7_+_120591170 | 0.22 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family, member 3 |
| chr13_-_30951282 | 0.22 |
ENST00000420219.1
|
LINC00426
|
long intergenic non-protein coding RNA 426 |
| chrX_-_54069253 | 0.21 |
ENST00000425862.1
ENST00000433120.1 |
PHF8
|
PHD finger protein 8 |
| chr6_+_28092338 | 0.20 |
ENST00000340487.4
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr11_-_8892464 | 0.19 |
ENST00000527347.1
ENST00000526241.1 ENST00000526126.1 ENST00000530938.1 ENST00000526057.1 |
ST5
|
suppression of tumorigenicity 5 |
| chrX_+_102611373 | 0.19 |
ENST00000372661.3
ENST00000372656.3 |
WBP5
|
WW domain binding protein 5 |
| chr10_+_180643 | 0.19 |
ENST00000509513.2
ENST00000397959.3 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr17_+_34171081 | 0.18 |
ENST00000585577.1
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chrX_+_49832231 | 0.18 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr6_-_52859046 | 0.18 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr11_+_28724129 | 0.18 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr16_-_53737722 | 0.18 |
ENST00000569716.1
ENST00000562588.1 ENST00000562230.1 ENST00000379925.3 ENST00000563746.1 ENST00000568653.3 |
RPGRIP1L
|
RPGRIP1-like |
| chr4_-_157892055 | 0.17 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr17_+_4675175 | 0.17 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr2_-_61389168 | 0.17 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr3_-_33686925 | 0.17 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr9_+_2159850 | 0.16 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_+_107364769 | 0.16 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr19_+_44617511 | 0.16 |
ENST00000262894.6
ENST00000588926.1 ENST00000592780.1 |
ZNF225
|
zinc finger protein 225 |
| chr3_-_149688971 | 0.16 |
ENST00000498307.1
ENST00000489155.1 |
PFN2
|
profilin 2 |
| chr11_-_82745238 | 0.15 |
ENST00000531021.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr2_-_69180083 | 0.15 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr9_-_85882145 | 0.15 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chrX_-_73512177 | 0.15 |
ENST00000603672.1
ENST00000418855.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr16_+_2533020 | 0.15 |
ENST00000562105.1
|
TBC1D24
|
TBC1 domain family, member 24 |
| chr4_+_56814968 | 0.15 |
ENST00000422247.2
|
CEP135
|
centrosomal protein 135kDa |
| chr19_-_44123734 | 0.15 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr5_-_74807418 | 0.15 |
ENST00000405807.4
ENST00000261415.7 |
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| chr5_+_79783788 | 0.14 |
ENST00000282226.4
|
FAM151B
|
family with sequence similarity 151, member B |
| chr3_-_158450475 | 0.14 |
ENST00000237696.5
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr11_-_85430356 | 0.14 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr7_-_139756791 | 0.14 |
ENST00000489809.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr4_-_56412713 | 0.14 |
ENST00000435527.2
|
CLOCK
|
clock circadian regulator |
| chr12_+_96337061 | 0.13 |
ENST00000266736.2
|
AMDHD1
|
amidohydrolase domain containing 1 |
| chr9_+_2158443 | 0.13 |
ENST00000302401.3
ENST00000324954.5 ENST00000423555.1 ENST00000382186.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_-_111291587 | 0.13 |
ENST00000437167.1
|
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr7_-_92146729 | 0.13 |
ENST00000541751.1
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr3_+_63898275 | 0.12 |
ENST00000538065.1
|
ATXN7
|
ataxin 7 |
| chr2_+_233404429 | 0.12 |
ENST00000389494.3
ENST00000389492.3 |
CHRNG
|
cholinergic receptor, nicotinic, gamma (muscle) |
| chr20_+_35504522 | 0.12 |
ENST00000602922.1
ENST00000217320.3 |
TLDC2
|
TBC/LysM-associated domain containing 2 |
| chr22_+_27068704 | 0.12 |
ENST00000444388.1
ENST00000450963.1 ENST00000449017.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr14_-_22005197 | 0.12 |
ENST00000541965.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr11_-_82782861 | 0.12 |
ENST00000524635.1
ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30
|
RAB30, member RAS oncogene family |
| chr12_-_15104040 | 0.12 |
ENST00000541644.1
ENST00000545895.1 |
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr7_+_23636992 | 0.12 |
ENST00000307471.3
ENST00000409765.1 |
CCDC126
|
coiled-coil domain containing 126 |
| chr7_-_151217166 | 0.12 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr7_+_151791074 | 0.12 |
ENST00000447796.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr8_-_74659693 | 0.11 |
ENST00000518767.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr3_+_99833755 | 0.11 |
ENST00000489081.1
|
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr2_-_153574480 | 0.11 |
ENST00000410080.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr22_+_31488433 | 0.11 |
ENST00000455608.1
|
SMTN
|
smoothelin |
| chr17_+_27573875 | 0.11 |
ENST00000225387.3
|
CRYBA1
|
crystallin, beta A1 |
| chr19_+_44488330 | 0.11 |
ENST00000591532.1
ENST00000407951.2 ENST00000270014.2 ENST00000590615.1 ENST00000586454.1 |
ZNF155
|
zinc finger protein 155 |
| chr15_+_67458861 | 0.11 |
ENST00000558428.1
ENST00000558827.1 |
SMAD3
|
SMAD family member 3 |
| chr2_-_61389240 | 0.11 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr12_-_113658826 | 0.11 |
ENST00000546692.1
|
IQCD
|
IQ motif containing D |
| chr7_-_14029283 | 0.11 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr11_-_125550726 | 0.11 |
ENST00000315608.3
ENST00000530048.1 |
ACRV1
|
acrosomal vesicle protein 1 |
| chr7_+_151791037 | 0.10 |
ENST00000419245.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr15_-_70994612 | 0.10 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr5_+_176784837 | 0.10 |
ENST00000408923.3
|
RGS14
|
regulator of G-protein signaling 14 |
| chr10_+_47894023 | 0.10 |
ENST00000358474.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr6_+_45296391 | 0.10 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr16_+_67360712 | 0.10 |
ENST00000569499.2
ENST00000329956.6 ENST00000561948.1 |
LRRC36
|
leucine rich repeat containing 36 |
| chr14_-_75083313 | 0.10 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr3_-_196910721 | 0.10 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr2_+_149447783 | 0.10 |
ENST00000449013.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr16_+_14280742 | 0.10 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr5_-_59783882 | 0.10 |
ENST00000505507.2
ENST00000502484.2 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr16_+_50308028 | 0.10 |
ENST00000566761.2
|
ADCY7
|
adenylate cyclase 7 |
| chr5_-_39203093 | 0.09 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr11_+_119056178 | 0.09 |
ENST00000525131.1
ENST00000531114.1 ENST00000355547.5 ENST00000322712.4 |
PDZD3
|
PDZ domain containing 3 |
| chr7_-_102985288 | 0.09 |
ENST00000379263.3
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr1_+_211813019 | 0.09 |
ENST00000430123.1
|
RP11-354K1.1
|
RP11-354K1.1 |
| chr6_+_146056706 | 0.09 |
ENST00000603994.1
|
RP3-466P17.1
|
RP3-466P17.1 |
| chr12_+_510742 | 0.09 |
ENST00000239830.4
|
CCDC77
|
coiled-coil domain containing 77 |
| chr1_+_104104379 | 0.09 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr4_-_144826682 | 0.09 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr5_+_148206156 | 0.09 |
ENST00000305988.4
|
ADRB2
|
adrenoceptor beta 2, surface |
| chrX_+_53078465 | 0.09 |
ENST00000375466.2
|
GPR173
|
G protein-coupled receptor 173 |
| chr2_+_234104079 | 0.09 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr22_-_42322795 | 0.09 |
ENST00000291232.3
|
TNFRSF13C
|
tumor necrosis factor receptor superfamily, member 13C |
| chr5_+_15500280 | 0.09 |
ENST00000504595.1
|
FBXL7
|
F-box and leucine-rich repeat protein 7 |
| chr12_-_112614506 | 0.09 |
ENST00000548588.2
|
HECTD4
|
HECT domain containing E3 ubiquitin protein ligase 4 |
| chr4_-_89744365 | 0.09 |
ENST00000513837.1
ENST00000503556.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr11_-_11374904 | 0.09 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr19_+_41281282 | 0.09 |
ENST00000263369.3
|
MIA
|
melanoma inhibitory activity |
| chr8_+_91233711 | 0.08 |
ENST00000523283.1
ENST00000517400.1 |
LINC00534
|
long intergenic non-protein coding RNA 534 |
| chr22_-_36236265 | 0.08 |
ENST00000414461.2
ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr19_+_41281060 | 0.08 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr14_-_35591679 | 0.08 |
ENST00000557278.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr12_+_32638897 | 0.08 |
ENST00000531134.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr12_-_95510743 | 0.08 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr10_+_24498060 | 0.08 |
ENST00000376454.3
ENST00000376452.3 |
KIAA1217
|
KIAA1217 |
| chr4_+_56815102 | 0.08 |
ENST00000257287.4
|
CEP135
|
centrosomal protein 135kDa |
| chr6_-_114292284 | 0.08 |
ENST00000520895.1
ENST00000521163.1 ENST00000524334.1 ENST00000368632.2 ENST00000398283.2 |
HDAC2
|
histone deacetylase 2 |
| chr16_-_53737795 | 0.08 |
ENST00000262135.4
ENST00000564374.1 ENST00000566096.1 |
RPGRIP1L
|
RPGRIP1-like |
| chr16_-_67360662 | 0.08 |
ENST00000304372.5
|
KCTD19
|
potassium channel tetramerization domain containing 19 |
| chr8_+_118532937 | 0.08 |
ENST00000297347.3
|
MED30
|
mediator complex subunit 30 |
| chr1_-_48866517 | 0.08 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr19_+_47538560 | 0.08 |
ENST00000439365.2
ENST00000594670.1 |
NPAS1
|
neuronal PAS domain protein 1 |
| chr2_-_153573887 | 0.08 |
ENST00000493468.2
ENST00000545856.1 |
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr15_-_78358465 | 0.08 |
ENST00000435468.1
|
TBC1D2B
|
TBC1 domain family, member 2B |
| chr12_+_51347705 | 0.08 |
ENST00000398455.3
|
HIGD1C
|
HIG1 hypoxia inducible domain family, member 1C |
| chr12_-_21910775 | 0.08 |
ENST00000539782.1
|
LDHB
|
lactate dehydrogenase B |
| chr9_-_117150243 | 0.08 |
ENST00000374088.3
|
AKNA
|
AT-hook transcription factor |
| chr2_+_105654441 | 0.08 |
ENST00000258455.3
|
MRPS9
|
mitochondrial ribosomal protein S9 |
| chr6_+_80816342 | 0.08 |
ENST00000369760.4
ENST00000356489.5 ENST00000320393.6 |
BCKDHB
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr10_-_75910539 | 0.08 |
ENST00000372745.1
|
AP3M1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr10_-_65028817 | 0.08 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr2_-_160472952 | 0.08 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr7_+_142919130 | 0.08 |
ENST00000408947.3
|
TAS2R40
|
taste receptor, type 2, member 40 |
| chr7_-_102985035 | 0.08 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr14_-_50474238 | 0.08 |
ENST00000399206.1
|
C14orf182
|
chromosome 14 open reading frame 182 |
| chr11_-_125550764 | 0.08 |
ENST00000527795.1
|
ACRV1
|
acrosomal vesicle protein 1 |
| chr6_-_41040268 | 0.08 |
ENST00000373154.2
ENST00000244558.9 ENST00000464633.1 ENST00000424266.2 ENST00000479950.1 ENST00000482515.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr10_+_180405 | 0.08 |
ENST00000439456.1
ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr1_+_236557569 | 0.08 |
ENST00000334232.4
|
EDARADD
|
EDAR-associated death domain |
| chr3_+_107364683 | 0.08 |
ENST00000413213.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chrX_-_41782683 | 0.07 |
ENST00000378163.1
ENST00000378154.1 |
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr1_-_54879140 | 0.07 |
ENST00000525990.1
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr16_+_12059091 | 0.07 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr19_+_36393367 | 0.07 |
ENST00000246551.4
|
HCST
|
hematopoietic cell signal transducer |
| chr14_-_37642016 | 0.07 |
ENST00000331299.5
|
SLC25A21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr3_-_188665428 | 0.07 |
ENST00000444488.1
|
TPRG1-AS1
|
TPRG1 antisense RNA 1 |
| chr1_+_36772691 | 0.07 |
ENST00000312808.4
ENST00000505871.1 |
SH3D21
|
SH3 domain containing 21 |
| chr10_-_65028938 | 0.07 |
ENST00000402544.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr6_+_143771934 | 0.07 |
ENST00000367592.1
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chr11_-_104840093 | 0.07 |
ENST00000417440.2
ENST00000444739.2 |
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr2_-_160473114 | 0.07 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr12_+_510795 | 0.07 |
ENST00000412006.2
|
CCDC77
|
coiled-coil domain containing 77 |
| chr13_-_27745936 | 0.07 |
ENST00000282344.6
|
USP12
|
ubiquitin specific peptidase 12 |
| chr9_-_75695323 | 0.07 |
ENST00000419959.1
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr7_-_17500294 | 0.07 |
ENST00000439046.1
|
AC019117.2
|
AC019117.2 |
| chr10_-_129924468 | 0.07 |
ENST00000368653.3
|
MKI67
|
marker of proliferation Ki-67 |
| chr6_-_41040195 | 0.07 |
ENST00000463088.1
ENST00000469104.1 ENST00000486443.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr4_-_185655212 | 0.07 |
ENST00000541971.1
|
MLF1IP
|
centromere protein U |
| chr10_-_13523073 | 0.07 |
ENST00000440282.1
|
BEND7
|
BEN domain containing 7 |
| chr19_-_49140609 | 0.07 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr17_+_3540043 | 0.07 |
ENST00000574218.1
|
CTNS
|
cystinosin, lysosomal cystine transporter |
| chr1_+_23695680 | 0.07 |
ENST00000454117.1
ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213
|
chromosome 1 open reading frame 213 |
| chr1_-_242612726 | 0.07 |
ENST00000459864.1
|
PLD5
|
phospholipase D family, member 5 |
| chr21_+_35014783 | 0.07 |
ENST00000381291.4
ENST00000381285.4 ENST00000399367.3 ENST00000399352.1 ENST00000399355.2 ENST00000399349.1 |
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr4_+_108852697 | 0.07 |
ENST00000508453.1
|
CYP2U1
|
cytochrome P450, family 2, subfamily U, polypeptide 1 |
| chr16_+_2014941 | 0.07 |
ENST00000531523.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr9_+_70856397 | 0.07 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr18_-_48723690 | 0.07 |
ENST00000406189.3
|
MEX3C
|
mex-3 RNA binding family member C |
| chr8_+_40010989 | 0.07 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr22_-_32334403 | 0.07 |
ENST00000543051.1
|
C22orf24
|
chromosome 22 open reading frame 24 |
| chr17_+_79495397 | 0.07 |
ENST00000417245.2
ENST00000334850.7 |
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr19_-_49140692 | 0.07 |
ENST00000222122.5
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr12_-_113658892 | 0.07 |
ENST00000299732.2
ENST00000416617.2 |
IQCD
|
IQ motif containing D |
| chrX_+_100353153 | 0.07 |
ENST00000423383.1
ENST00000218507.5 ENST00000403304.2 ENST00000435570.1 |
CENPI
|
centromere protein I |
| chr5_+_147763498 | 0.07 |
ENST00000340253.5
|
FBXO38
|
F-box protein 38 |
| chr14_-_95623607 | 0.07 |
ENST00000531162.1
ENST00000529720.1 ENST00000343455.3 |
DICER1
|
dicer 1, ribonuclease type III |
| chr17_+_1665345 | 0.07 |
ENST00000576406.1
ENST00000571149.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr4_-_144940477 | 0.07 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr17_+_76311791 | 0.07 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr12_-_50419177 | 0.07 |
ENST00000454520.2
ENST00000546595.1 ENST00000548824.1 ENST00000549777.1 ENST00000546723.1 ENST00000427314.2 ENST00000552157.1 ENST00000552310.1 ENST00000548644.1 ENST00000312377.5 ENST00000546786.1 ENST00000550149.1 ENST00000546764.1 ENST00000552004.1 ENST00000548320.1 ENST00000547905.1 ENST00000550651.1 ENST00000551145.1 ENST00000434422.1 ENST00000552921.1 |
RACGAP1
|
Rac GTPase activating protein 1 |
| chrX_+_17755696 | 0.07 |
ENST00000419185.1
|
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr7_-_105332084 | 0.07 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr2_+_153574428 | 0.07 |
ENST00000326446.5
|
ARL6IP6
|
ADP-ribosylation-like factor 6 interacting protein 6 |
| chr4_+_41614720 | 0.06 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr21_+_35014706 | 0.06 |
ENST00000399353.1
ENST00000444491.1 ENST00000381318.3 |
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr20_+_16729003 | 0.06 |
ENST00000246081.2
|
OTOR
|
otoraplin |
| chr17_-_41836153 | 0.06 |
ENST00000301691.2
|
SOST
|
sclerostin |
| chr4_-_89951028 | 0.06 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr1_+_164528437 | 0.06 |
ENST00000485769.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr6_-_114292449 | 0.06 |
ENST00000519065.1
|
HDAC2
|
histone deacetylase 2 |
| chr1_+_164529004 | 0.06 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr2_+_234601512 | 0.06 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr12_-_58212487 | 0.06 |
ENST00000549994.1
|
AVIL
|
advillin |
| chr11_+_107799118 | 0.06 |
ENST00000320578.2
|
RAB39A
|
RAB39A, member RAS oncogene family |
| chr9_+_36190905 | 0.06 |
ENST00000345519.5
ENST00000470744.1 ENST00000242285.6 ENST00000466396.1 ENST00000396603.2 |
CLTA
|
clathrin, light chain A |
| chr3_-_33686743 | 0.06 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr4_+_41362796 | 0.06 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_+_53068020 | 0.06 |
ENST00000361314.4
|
GPX7
|
glutathione peroxidase 7 |
| chr3_-_110612059 | 0.06 |
ENST00000485473.1
|
RP11-553A10.1
|
Uncharacterized protein |
| chr4_-_185654511 | 0.06 |
ENST00000514781.1
|
MLF1IP
|
centromere protein U |
| chrX_-_85302531 | 0.06 |
ENST00000537751.1
ENST00000358786.4 ENST00000357749.2 |
CHM
|
choroideremia (Rab escort protein 1) |
| chr4_+_9446156 | 0.06 |
ENST00000334879.1
|
DEFB131
|
defensin, beta 131 |
| chr9_-_117150303 | 0.06 |
ENST00000312033.3
|
AKNA
|
AT-hook transcription factor |
| chr5_+_59783540 | 0.06 |
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr22_-_36236623 | 0.06 |
ENST00000405409.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr5_+_55354822 | 0.06 |
ENST00000511861.1
|
CTD-2227I18.1
|
CTD-2227I18.1 |
| chr11_-_10920714 | 0.06 |
ENST00000533941.1
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr21_+_35014829 | 0.06 |
ENST00000451686.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr17_+_38337491 | 0.06 |
ENST00000538981.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr18_+_21452804 | 0.06 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr2_+_172544182 | 0.06 |
ENST00000409197.1
ENST00000456808.1 ENST00000409317.1 ENST00000409773.1 ENST00000411953.1 ENST00000409453.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr17_+_75450099 | 0.06 |
ENST00000586433.1
|
SEPT9
|
septin 9 |
| chr6_+_90192974 | 0.06 |
ENST00000520458.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr10_-_129924611 | 0.06 |
ENST00000368654.3
|
MKI67
|
marker of proliferation Ki-67 |
| chr20_+_45338126 | 0.06 |
ENST00000359271.2
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr1_+_61542922 | 0.06 |
ENST00000407417.3
|
NFIA
|
nuclear factor I/A |
| chr10_-_75910789 | 0.06 |
ENST00000355264.4
|
AP3M1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr21_+_17214724 | 0.06 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr12_+_121570631 | 0.06 |
ENST00000546057.1
ENST00000377162.2 ENST00000328963.5 ENST00000535250.1 ENST00000541446.1 |
P2RX7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GFI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.2 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.2 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:0033488 | cholesterol biosynthetic process via 24,25-dihydrolanosterol(GO:0033488) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.0 | 0.1 | GO:0043132 | NAD transport(GO:0043132) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.2 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.0 | GO:1901656 | cellular response to mycotoxin(GO:0036146) glycoside transport(GO:1901656) cellular response to bile acid(GO:1903413) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.0 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.0 | GO:1901147 | pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) thyroid-stimulating hormone secretion(GO:0070460) kidney field specification(GO:0072004) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.0 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.0 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0008398 | sterol 14-demethylase activity(GO:0008398) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.0 | GO:0031775 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.0 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.0 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.0 | 0.1 | GO:0030249 | cyclase regulator activity(GO:0010851) guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |