Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
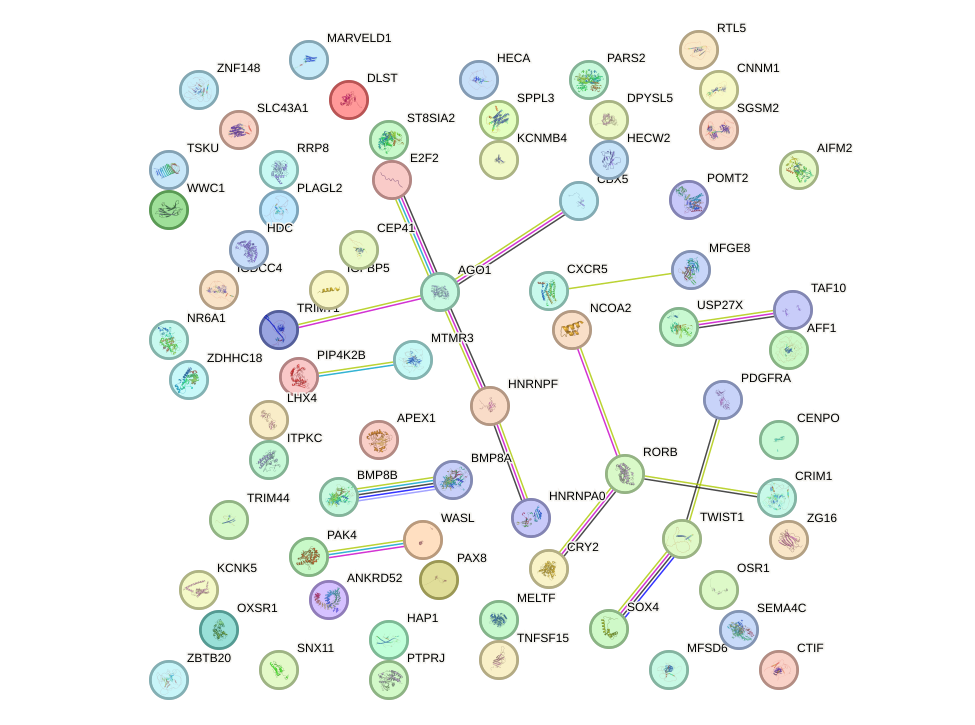
Results for GCCUGUC
Z-value: 0.08
miRNA associated with seed GCCUGUC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-214-5p
|
MIMAT0004564 |
Activity profile of GCCUGUC motif
Sorted Z-values of GCCUGUC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_118754475 | 0.03 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr12_-_56652111 | 0.02 |
ENST00000267116.7
|
ANKRD52
|
ankyrin repeat domain 52 |
| chr11_+_76494253 | 0.01 |
ENST00000333090.4
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr5_-_137090028 | 0.01 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr17_+_46184911 | 0.01 |
ENST00000580219.1
ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11
|
sorting nexin 11 |
| chr7_-_19157248 | 0.01 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1 |
| chr2_-_97535708 | 0.01 |
ENST00000305476.5
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr1_+_39957292 | 0.01 |
ENST00000331593.5
|
BMP8A
|
bone morphogenetic protein 8a |
| chr1_+_27153173 | 0.01 |
ENST00000374142.4
|
ZDHHC18
|
zinc finger, DHHC-type containing 18 |
| chr3_-_114790179 | 0.01 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr6_-_39197226 | 0.01 |
ENST00000359534.3
|
KCNK5
|
potassium channel, subfamily K, member 5 |
| chr6_+_21593972 | 0.01 |
ENST00000244745.1
ENST00000543472.1 |
SOX4
|
SRY (sex determining region Y)-box 4 |
| chr11_-_6633799 | 0.01 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr22_-_50746027 | 0.01 |
ENST00000425954.1
ENST00000449103.1 |
PLXNB2
|
plexin B2 |
| chr4_-_39460520 | 0.01 |
ENST00000503040.1
ENST00000504470.1 ENST00000508595.1 ENST00000295955.9 |
RPL9
|
ribosomal protein L9 |
| chr11_-_57283159 | 0.01 |
ENST00000533263.1
ENST00000278426.3 |
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr11_+_45868957 | 0.01 |
ENST00000443527.2
|
CRY2
|
cryptochrome 2 (photolyase-like) |
| chr11_-_6624801 | 0.01 |
ENST00000534343.1
ENST00000254605.6 |
RRP8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr12_-_54673871 | 0.01 |
ENST00000209875.4
|
CBX5
|
chromobox homolog 5 |
| chr1_-_23857698 | 0.00 |
ENST00000361729.2
|
E2F2
|
E2F transcription factor 2 |
| chr10_+_99473455 | 0.00 |
ENST00000285605.6
|
MARVELD1
|
MARVEL domain containing 1 |
| chr22_+_30279144 | 0.00 |
ENST00000401950.2
ENST00000333027.3 ENST00000445401.1 ENST00000323630.5 ENST00000351488.3 |
MTMR3
|
myotubularin related protein 3 |
| chr19_+_41222998 | 0.00 |
ENST00000263370.2
|
ITPKC
|
inositol-trisphosphate 3-kinase C |
| chr2_+_25015968 | 0.00 |
ENST00000380834.2
ENST00000473706.1 |
CENPO
|
centromere protein O |
| chr9_-_127533519 | 0.00 |
ENST00000487099.2
ENST00000344523.4 ENST00000373584.3 |
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr5_+_95187934 | 0.00 |
ENST00000357880.3
ENST00000436592.1 |
C5orf27
|
chromosome 5 open reading frame 27 |
| chr2_-_19558373 | 0.00 |
ENST00000272223.2
|
OSR1
|
odd-skipped related transciption factor 1 |
| chr11_+_35684288 | 0.00 |
ENST00000299413.5
|
TRIM44
|
tripartite motif containing 44 |
| chr10_-_71892555 | 0.00 |
ENST00000307864.1
|
AIFM2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr1_+_36348790 | 0.00 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr12_-_121342170 | 0.00 |
ENST00000353487.2
|
SPPL3
|
signal peptide peptidase like 3 |
| chr6_+_44187242 | 0.00 |
ENST00000393844.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr17_-_61523535 | 0.00 |
ENST00000584031.1
ENST00000392976.1 |
CYB561
|
cytochrome b561 |
| chr17_+_2240775 | 0.00 |
ENST00000268989.3
ENST00000426855.2 |
SGSM2
|
small G protein signaling modulator 2 |
| chr19_-_15560730 | 0.00 |
ENST00000389282.4
ENST00000263381.7 |
WIZ
|
widely interspaced zinc finger motifs |
| chr15_-_65715401 | 0.00 |
ENST00000352385.2
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr18_+_46065393 | 0.00 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr2_+_191273052 | 0.00 |
ENST00000417958.1
ENST00000432036.1 ENST00000392328.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr1_+_180199393 | 0.00 |
ENST00000263726.2
|
LHX4
|
LIM homeobox 4 |
| chr11_-_118305921 | 0.00 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr16_+_29789561 | 0.00 |
ENST00000400752.4
|
ZG16
|
zymogen granule protein 16 |
| chr19_+_39616410 | 0.00 |
ENST00000602004.1
ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr20_-_30795511 | 0.00 |
ENST00000246229.4
|
PLAGL2
|
pleiomorphic adenoma gene-like 2 |
| chr3_+_32859510 | 0.00 |
ENST00000383763.5
|
TRIM71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr10_+_101088836 | 0.00 |
ENST00000356713.4
|
CNNM1
|
cyclin M1 |
| chrX_-_71351678 | 0.00 |
ENST00000609883.1
ENST00000545866.1 |
RGAG4
|
retrotransposon gag domain containing 4 |
| chr14_+_75348592 | 0.00 |
ENST00000334220.4
|
DLST
|
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| chr3_-_196756646 | 0.00 |
ENST00000439320.1
ENST00000296351.4 ENST00000296350.5 |
MFI2
|
antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 |
| chr15_-_41408409 | 0.00 |
ENST00000361937.3
|
INO80
|
INO80 complex subunit |
| chr15_-_89456593 | 0.00 |
ENST00000558029.1
ENST00000539437.1 ENST00000542878.1 ENST00000268151.7 ENST00000566497.1 |
MFGE8
|
milk fat globule-EGF factor 8 protein |
| chr2_+_134877740 | 0.00 |
ENST00000409645.1
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr9_+_77112244 | 0.00 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr14_-_77787198 | 0.00 |
ENST00000261534.4
|
POMT2
|
protein-O-mannosyltransferase 2 |
| chr4_+_87856129 | 0.00 |
ENST00000395146.4
ENST00000507468.1 |
AFF1
|
AF4/FMR2 family, member 1 |
| chr15_-_50558223 | 0.00 |
ENST00000267845.3
|
HDC
|
histidine decarboxylase |
| chr1_-_55230165 | 0.00 |
ENST00000371279.3
|
PARS2
|
prolyl-tRNA synthetase 2, mitochondrial (putative) |
| chr9_-_74980113 | 0.00 |
ENST00000376962.5
ENST00000376960.4 ENST00000237937.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr15_+_92937058 | 0.00 |
ENST00000268164.3
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr10_-_43903217 | 0.00 |
ENST00000357065.4
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr17_-_39890893 | 0.00 |
ENST00000393939.2
ENST00000347901.4 ENST00000341193.5 ENST00000310778.5 |
HAP1
|
huntingtin-associated protein 1 |
| chr2_-_114036488 | 0.00 |
ENST00000263335.7
ENST00000397647.3 ENST00000348715.5 ENST00000429538.3 |
PAX8
|
paired box 8 |
| chr2_-_197457335 | 0.00 |
ENST00000260983.3
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr11_+_48002076 | 0.00 |
ENST00000418331.2
ENST00000440289.2 |
PTPRJ
|
protein tyrosine phosphatase, receptor type, J |
| chr9_-_117568365 | 0.00 |
ENST00000374045.4
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15 |
| chr2_+_36582857 | 0.00 |
ENST00000280527.2
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
| chr12_+_70760056 | 0.00 |
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chrX_+_49644470 | 0.00 |
ENST00000508866.2
|
USP27X
|
ubiquitin specific peptidase 27, X-linked |
| chr2_+_27070964 | 0.00 |
ENST00000288699.6
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr7_-_130080977 | 0.00 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr8_-_71316021 | 0.00 |
ENST00000452400.2
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr3_-_125094093 | 0.00 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr2_-_217560248 | 0.00 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr7_-_123389104 | 0.00 |
ENST00000223023.4
|
WASL
|
Wiskott-Aldrich syndrome-like |
| chr5_+_167718604 | 0.00 |
ENST00000265293.4
|
WWC1
|
WW and C2 domain containing 1 |
| chr4_+_55095264 | 0.00 |
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr17_-_36956155 | 0.00 |
ENST00000269554.3
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta |