Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
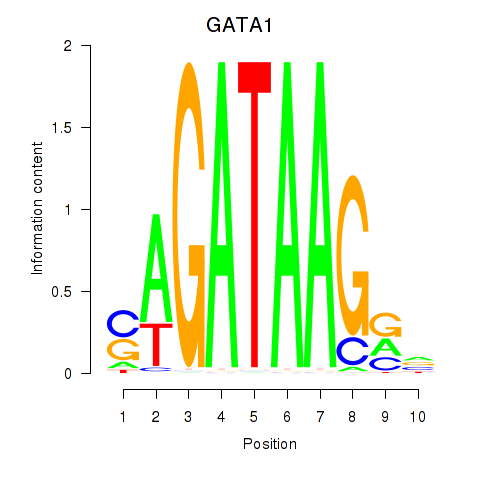
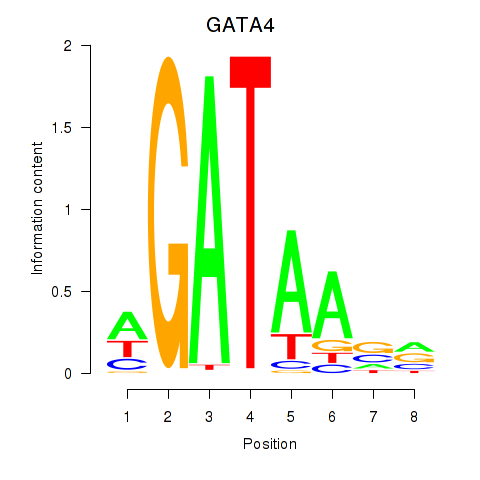
Results for GATA1_GATA4
Z-value: 1.30
Transcription factors associated with GATA1_GATA4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA1
|
ENSG00000102145.9 | GATA binding protein 1 |
|
GATA4
|
ENSG00000136574.13 | GATA binding protein 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA4 | hg19_v2_chr8_+_11534462_11534475 | 0.60 | 4.0e-01 | Click! |
| GATA1 | hg19_v2_chrX_+_48644962_48644983 | -0.35 | 6.5e-01 | Click! |
Activity profile of GATA1_GATA4 motif
Sorted Z-values of GATA1_GATA4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_42630389 | 1.25 |
ENST00000357001.2
|
GUCA2A
|
guanylate cyclase activator 2A (guanylin) |
| chr1_-_226187013 | 0.83 |
ENST00000272091.7
|
SDE2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr1_-_238108575 | 0.72 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr11_-_10920838 | 0.68 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr10_-_71993176 | 0.67 |
ENST00000373232.3
|
PPA1
|
pyrophosphatase (inorganic) 1 |
| chr17_-_79817091 | 0.66 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr11_-_61734599 | 0.62 |
ENST00000532601.1
|
FTH1
|
ferritin, heavy polypeptide 1 |
| chr6_+_31691121 | 0.59 |
ENST00000480039.1
ENST00000375810.4 ENST00000375805.2 ENST00000375809.3 ENST00000375804.2 ENST00000375814.3 ENST00000375806.2 |
C6orf25
|
chromosome 6 open reading frame 25 |
| chr19_-_55669093 | 0.57 |
ENST00000344887.5
|
TNNI3
|
troponin I type 3 (cardiac) |
| chr19_-_39303576 | 0.56 |
ENST00000594209.1
|
LGALS4
|
lectin, galactoside-binding, soluble, 4 |
| chr10_+_28822636 | 0.51 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr19_+_37808831 | 0.51 |
ENST00000589801.1
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr18_-_8337038 | 0.50 |
ENST00000594251.1
|
AP001094.1
|
Uncharacterized protein |
| chr2_+_189156389 | 0.48 |
ENST00000409843.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr15_+_49715293 | 0.48 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr8_+_124194875 | 0.47 |
ENST00000522648.1
ENST00000276699.6 |
FAM83A
|
family with sequence similarity 83, member A |
| chr4_+_74606223 | 0.46 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr11_-_85393886 | 0.46 |
ENST00000534224.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr7_-_99277610 | 0.45 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr21_-_43786634 | 0.44 |
ENST00000291527.2
|
TFF1
|
trefoil factor 1 |
| chrX_-_100129320 | 0.43 |
ENST00000372966.3
|
NOX1
|
NADPH oxidase 1 |
| chr2_+_192543694 | 0.43 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr14_+_105953204 | 0.41 |
ENST00000409393.2
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr10_+_180643 | 0.41 |
ENST00000509513.2
ENST00000397959.3 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr15_+_49715449 | 0.40 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr12_-_30907862 | 0.39 |
ENST00000541765.1
ENST00000537108.1 |
CAPRIN2
|
caprin family member 2 |
| chr12_-_30907822 | 0.38 |
ENST00000540436.1
|
CAPRIN2
|
caprin family member 2 |
| chr10_+_71075516 | 0.37 |
ENST00000436817.1
|
HK1
|
hexokinase 1 |
| chr6_-_10115007 | 0.36 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr12_+_104337515 | 0.36 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr16_+_333152 | 0.36 |
ENST00000219406.6
ENST00000404312.1 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chr2_-_44065889 | 0.35 |
ENST00000543989.1
ENST00000405322.1 |
ABCG5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr14_+_102276209 | 0.33 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr12_+_4430371 | 0.33 |
ENST00000179259.4
|
C12orf5
|
chromosome 12 open reading frame 5 |
| chr11_+_63974135 | 0.33 |
ENST00000544997.1
ENST00000345728.5 ENST00000279227.5 |
FERMT3
|
fermitin family member 3 |
| chr6_+_30908747 | 0.32 |
ENST00000462446.1
ENST00000304311.2 |
DPCR1
|
diffuse panbronchiolitis critical region 1 |
| chr4_+_165675197 | 0.32 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr3_-_119379427 | 0.32 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr19_+_11650709 | 0.32 |
ENST00000586059.1
|
CNN1
|
calponin 1, basic, smooth muscle |
| chr2_-_74555350 | 0.31 |
ENST00000444570.1
|
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr9_-_114937465 | 0.31 |
ENST00000355396.3
|
SUSD1
|
sushi domain containing 1 |
| chr8_+_124194752 | 0.31 |
ENST00000318462.6
|
FAM83A
|
family with sequence similarity 83, member A |
| chr12_-_30907749 | 0.30 |
ENST00000542550.1
ENST00000540584.1 |
CAPRIN2
|
caprin family member 2 |
| chr10_+_28822417 | 0.30 |
ENST00000428935.1
ENST00000420266.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chrX_+_102965835 | 0.29 |
ENST00000319560.6
|
TMEM31
|
transmembrane protein 31 |
| chr7_-_1980128 | 0.29 |
ENST00000437877.1
|
MAD1L1
|
MAD1 mitotic arrest deficient-like 1 (yeast) |
| chr11_-_125648690 | 0.28 |
ENST00000436890.2
ENST00000358524.3 |
PATE2
|
prostate and testis expressed 2 |
| chr16_+_85936295 | 0.28 |
ENST00000563180.1
ENST00000564617.1 ENST00000564803.1 |
IRF8
|
interferon regulatory factor 8 |
| chr4_-_52883786 | 0.28 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr1_-_11918988 | 0.27 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr4_-_103266219 | 0.27 |
ENST00000394833.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr6_+_27356745 | 0.27 |
ENST00000461521.1
|
ZNF391
|
zinc finger protein 391 |
| chr4_+_76871883 | 0.26 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr3_-_57233966 | 0.26 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr10_+_180405 | 0.26 |
ENST00000439456.1
ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr2_+_109237717 | 0.26 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr11_-_57102947 | 0.26 |
ENST00000526696.1
|
SSRP1
|
structure specific recognition protein 1 |
| chr6_-_31846744 | 0.26 |
ENST00000414427.1
ENST00000229729.6 ENST00000375562.4 |
SLC44A4
|
solute carrier family 44, member 4 |
| chr12_-_56120838 | 0.26 |
ENST00000548160.1
|
CD63
|
CD63 molecule |
| chr12_-_105629852 | 0.24 |
ENST00000551662.1
ENST00000553097.1 |
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr9_-_114937543 | 0.24 |
ENST00000374264.2
ENST00000374263.3 |
SUSD1
|
sushi domain containing 1 |
| chr8_+_107630340 | 0.24 |
ENST00000497705.1
|
OXR1
|
oxidation resistance 1 |
| chr8_-_91618285 | 0.24 |
ENST00000517505.1
|
LINC01030
|
long intergenic non-protein coding RNA 1030 |
| chr2_+_189156586 | 0.24 |
ENST00000409830.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr9_+_90341024 | 0.24 |
ENST00000340342.6
ENST00000342020.5 |
CTSL
|
cathepsin L |
| chr16_+_81070792 | 0.23 |
ENST00000564241.1
ENST00000565237.1 |
ATMIN
|
ATM interactor |
| chr3_+_172468505 | 0.23 |
ENST00000427830.1
ENST00000417960.1 ENST00000428567.1 ENST00000366090.2 ENST00000426894.1 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr13_+_53029564 | 0.23 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr14_-_38036271 | 0.23 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr11_-_125550726 | 0.23 |
ENST00000315608.3
ENST00000530048.1 |
ACRV1
|
acrosomal vesicle protein 1 |
| chr9_+_86237963 | 0.23 |
ENST00000277124.8
|
IDNK
|
idnK, gluconokinase homolog (E. coli) |
| chr7_+_150549565 | 0.23 |
ENST00000360937.4
ENST00000416793.2 ENST00000483043.1 |
AOC1
|
amine oxidase, copper containing 1 |
| chr14_+_105952648 | 0.23 |
ENST00000330233.7
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr1_-_235098935 | 0.22 |
ENST00000423175.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr1_+_234509413 | 0.22 |
ENST00000366613.1
ENST00000366612.1 |
COA6
|
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chr9_-_88896977 | 0.22 |
ENST00000311534.6
|
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr9_+_33240157 | 0.22 |
ENST00000379721.3
|
SPINK4
|
serine peptidase inhibitor, Kazal type 4 |
| chr9_+_140125385 | 0.22 |
ENST00000361134.2
|
SLC34A3
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 3 |
| chr15_-_72462661 | 0.22 |
ENST00000570275.1
|
GRAMD2
|
GRAM domain containing 2 |
| chr10_+_70847852 | 0.21 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr13_-_46425865 | 0.21 |
ENST00000400405.2
|
SIAH3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr3_-_119379719 | 0.21 |
ENST00000493094.1
|
POPDC2
|
popeye domain containing 2 |
| chr12_-_121476750 | 0.21 |
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr10_+_28822236 | 0.21 |
ENST00000347934.4
ENST00000354911.4 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr9_+_132099158 | 0.21 |
ENST00000444125.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr16_+_70680439 | 0.20 |
ENST00000288098.2
|
IL34
|
interleukin 34 |
| chr6_-_30128657 | 0.20 |
ENST00000449742.2
ENST00000376704.3 |
TRIM10
|
tripartite motif containing 10 |
| chr20_-_60294804 | 0.20 |
ENST00000317652.1
|
RP11-429E11.3
|
Uncharacterized protein |
| chr22_-_43042955 | 0.20 |
ENST00000402438.1
|
CYB5R3
|
cytochrome b5 reductase 3 |
| chrX_+_135278908 | 0.20 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr16_+_85832146 | 0.20 |
ENST00000565078.1
|
COX4I1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr4_-_65275162 | 0.20 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr10_+_71075552 | 0.19 |
ENST00000298649.3
|
HK1
|
hexokinase 1 |
| chrX_-_43832711 | 0.19 |
ENST00000378062.5
|
NDP
|
Norrie disease (pseudoglioma) |
| chr10_-_101673782 | 0.19 |
ENST00000422692.1
|
DNMBP
|
dynamin binding protein |
| chr6_-_107235378 | 0.19 |
ENST00000606430.1
|
RP1-60O19.1
|
RP1-60O19.1 |
| chr4_+_154387480 | 0.19 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr8_-_143961236 | 0.19 |
ENST00000377675.3
ENST00000517471.1 ENST00000292427.4 |
CYP11B1
|
cytochrome P450, family 11, subfamily B, polypeptide 1 |
| chr8_+_121137333 | 0.19 |
ENST00000309791.4
ENST00000297848.3 ENST00000247781.3 |
COL14A1
|
collagen, type XIV, alpha 1 |
| chr12_-_21910775 | 0.19 |
ENST00000539782.1
|
LDHB
|
lactate dehydrogenase B |
| chr2_+_192543153 | 0.19 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr4_-_89619386 | 0.18 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr11_-_125550764 | 0.18 |
ENST00000527795.1
|
ACRV1
|
acrosomal vesicle protein 1 |
| chr10_-_96122682 | 0.18 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr2_+_197504278 | 0.18 |
ENST00000272831.7
ENST00000389175.4 ENST00000472405.2 ENST00000423093.2 |
CCDC150
|
coiled-coil domain containing 150 |
| chr3_-_135916073 | 0.18 |
ENST00000481989.1
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr3_-_58196939 | 0.18 |
ENST00000394549.2
ENST00000461914.3 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr19_+_17392672 | 0.18 |
ENST00000594072.1
ENST00000598347.1 |
ANKLE1
|
ankyrin repeat and LEM domain containing 1 |
| chr6_+_155537771 | 0.18 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr2_-_165424973 | 0.18 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr11_+_76571911 | 0.17 |
ENST00000534206.1
ENST00000532485.1 ENST00000526597.1 ENST00000533873.1 ENST00000538157.1 |
ACER3
|
alkaline ceramidase 3 |
| chr11_-_134123142 | 0.17 |
ENST00000392595.2
ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1
|
thymocyte nuclear protein 1 |
| chr5_+_66300464 | 0.17 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr5_+_81601166 | 0.17 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr12_-_105630016 | 0.17 |
ENST00000258530.3
|
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr2_+_189156721 | 0.17 |
ENST00000409927.1
ENST00000409805.1 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr15_+_78632666 | 0.17 |
ENST00000299529.6
|
CRABP1
|
cellular retinoic acid binding protein 1 |
| chr1_-_25747283 | 0.17 |
ENST00000346452.4
ENST00000340849.4 ENST00000349438.4 ENST00000294413.7 ENST00000413854.1 ENST00000455194.1 ENST00000243186.6 ENST00000425135.1 |
RHCE
|
Rh blood group, CcEe antigens |
| chr11_+_34642656 | 0.17 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr7_-_50860565 | 0.17 |
ENST00000403097.1
|
GRB10
|
growth factor receptor-bound protein 10 |
| chr5_+_67584523 | 0.17 |
ENST00000521409.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr12_+_75874984 | 0.16 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr6_+_126102292 | 0.16 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr19_-_12997995 | 0.16 |
ENST00000264834.4
|
KLF1
|
Kruppel-like factor 1 (erythroid) |
| chr1_-_225615599 | 0.16 |
ENST00000421383.1
ENST00000272163.4 |
LBR
|
lamin B receptor |
| chr4_-_144826682 | 0.16 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr16_+_14280564 | 0.16 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr1_-_24469602 | 0.16 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chrX_+_118892545 | 0.16 |
ENST00000343905.3
|
SOWAHD
|
sosondowah ankyrin repeat domain family member D |
| chr19_-_58485895 | 0.16 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr4_-_100815525 | 0.16 |
ENST00000226522.8
ENST00000499666.2 |
LAMTOR3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr13_-_96329048 | 0.16 |
ENST00000606011.1
ENST00000499499.2 |
DNAJC3-AS1
|
DNAJC3 antisense RNA 1 (head to head) |
| chr19_-_11849697 | 0.16 |
ENST00000586121.1
ENST00000431998.1 ENST00000341191.6 ENST00000545749.1 ENST00000440527.1 |
ZNF823
|
zinc finger protein 823 |
| chr17_-_57229155 | 0.16 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr11_-_105892937 | 0.16 |
ENST00000301919.4
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr6_+_33168189 | 0.16 |
ENST00000444757.1
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr6_-_107235331 | 0.16 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr17_+_75449889 | 0.16 |
ENST00000590938.1
|
SEPT9
|
septin 9 |
| chr12_-_56221330 | 0.15 |
ENST00000546837.1
|
RP11-762I7.5
|
Uncharacterized protein |
| chr16_-_67978016 | 0.15 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr1_-_154150651 | 0.15 |
ENST00000302206.5
|
TPM3
|
tropomyosin 3 |
| chr2_-_69180083 | 0.15 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr19_-_51568324 | 0.15 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr2_-_106054952 | 0.15 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr1_-_155270770 | 0.15 |
ENST00000392414.3
|
PKLR
|
pyruvate kinase, liver and RBC |
| chr11_-_47400062 | 0.15 |
ENST00000533030.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr21_-_36421401 | 0.15 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr3_-_197686847 | 0.15 |
ENST00000265239.6
|
IQCG
|
IQ motif containing G |
| chr20_-_271009 | 0.15 |
ENST00000382369.5
|
C20orf96
|
chromosome 20 open reading frame 96 |
| chr17_+_72920370 | 0.15 |
ENST00000331427.4
|
OTOP2
|
otopetrin 2 |
| chr12_+_13061894 | 0.15 |
ENST00000540125.1
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A |
| chr10_-_71993138 | 0.15 |
ENST00000608321.1
|
PPA1
|
pyrophosphatase (inorganic) 1 |
| chr14_-_53258180 | 0.15 |
ENST00000554230.1
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr12_+_69633317 | 0.15 |
ENST00000435070.2
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr4_-_130692631 | 0.15 |
ENST00000500092.2
ENST00000509105.1 |
RP11-519M16.1
|
RP11-519M16.1 |
| chr4_-_84035905 | 0.15 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr5_+_157602404 | 0.15 |
ENST00000522975.1
|
CTC-436K13.1
|
CTC-436K13.1 |
| chr11_-_64510409 | 0.15 |
ENST00000394429.1
ENST00000394428.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr1_-_154164534 | 0.15 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr7_+_94536514 | 0.15 |
ENST00000413325.1
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr11_-_6008215 | 0.15 |
ENST00000332249.4
|
OR52L1
|
olfactory receptor, family 52, subfamily L, member 1 |
| chr7_-_99381884 | 0.14 |
ENST00000336411.2
|
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr1_-_161207986 | 0.14 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr4_+_81118647 | 0.14 |
ENST00000415738.2
|
PRDM8
|
PR domain containing 8 |
| chr21_-_34915084 | 0.14 |
ENST00000426819.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr2_+_201242715 | 0.14 |
ENST00000421573.1
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr4_+_75174204 | 0.14 |
ENST00000332112.4
ENST00000514968.1 ENST00000503098.1 ENST00000502358.1 ENST00000509145.1 ENST00000505212.1 |
EPGN
|
epithelial mitogen |
| chrX_+_84258832 | 0.14 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr13_-_31736478 | 0.14 |
ENST00000445273.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr6_-_138833630 | 0.14 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr3_-_145940126 | 0.14 |
ENST00000498625.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chrX_+_38211777 | 0.14 |
ENST00000039007.4
|
OTC
|
ornithine carbamoyltransferase |
| chr14_-_81893734 | 0.14 |
ENST00000555447.1
|
STON2
|
stonin 2 |
| chr6_-_150039249 | 0.14 |
ENST00000543571.1
|
LATS1
|
large tumor suppressor kinase 1 |
| chr17_-_38938786 | 0.13 |
ENST00000301656.3
|
KRT27
|
keratin 27 |
| chrX_+_135279179 | 0.13 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr17_+_75450099 | 0.13 |
ENST00000586433.1
|
SEPT9
|
septin 9 |
| chr14_-_21945057 | 0.13 |
ENST00000397762.1
|
RAB2B
|
RAB2B, member RAS oncogene family |
| chr8_+_41347915 | 0.13 |
ENST00000518270.1
ENST00000520817.1 |
GOLGA7
|
golgin A7 |
| chr4_+_99916765 | 0.13 |
ENST00000296411.6
|
METAP1
|
methionyl aminopeptidase 1 |
| chr19_-_36822551 | 0.13 |
ENST00000591372.1
|
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chr3_-_195310802 | 0.13 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr13_-_107214291 | 0.13 |
ENST00000375926.1
|
ARGLU1
|
arginine and glutamate rich 1 |
| chr8_-_41655107 | 0.13 |
ENST00000347528.4
ENST00000289734.7 ENST00000379758.2 ENST00000396945.1 ENST00000396942.1 ENST00000352337.4 |
ANK1
|
ankyrin 1, erythrocytic |
| chr14_-_45603657 | 0.13 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr3_-_58196688 | 0.13 |
ENST00000486455.1
|
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr7_+_128379346 | 0.13 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr12_+_51347705 | 0.13 |
ENST00000398455.3
|
HIGD1C
|
HIG1 hypoxia inducible domain family, member 1C |
| chr9_-_114937676 | 0.13 |
ENST00000374270.3
|
SUSD1
|
sushi domain containing 1 |
| chr10_+_31610064 | 0.13 |
ENST00000446923.2
ENST00000559476.1 |
ZEB1
|
zinc finger E-box binding homeobox 1 |
| chr15_-_52861394 | 0.13 |
ENST00000563277.1
ENST00000566423.1 |
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa |
| chr2_+_106361333 | 0.13 |
ENST00000233154.4
ENST00000451463.2 |
NCK2
|
NCK adaptor protein 2 |
| chr12_-_91546926 | 0.13 |
ENST00000550758.1
|
DCN
|
decorin |
| chr17_-_71258019 | 0.12 |
ENST00000344935.4
|
CPSF4L
|
cleavage and polyadenylation specific factor 4-like |
| chr6_+_14117872 | 0.12 |
ENST00000379153.3
|
CD83
|
CD83 molecule |
| chr5_+_169011033 | 0.12 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr7_-_107880508 | 0.12 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr1_+_150954493 | 0.12 |
ENST00000368947.4
|
ANXA9
|
annexin A9 |
| chr17_+_70036164 | 0.12 |
ENST00000602013.1
|
AC007461.1
|
Uncharacterized protein |
| chr7_-_100239132 | 0.12 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA1_GATA4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.2 | 0.9 | GO:0060437 | lung growth(GO:0060437) |
| 0.2 | 0.8 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.1 | 0.6 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 0.6 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.4 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.1 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.1 | 1.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.1 | 0.3 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.2 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal lipid absorption(GO:1904729) |
| 0.1 | 0.3 | GO:0044026 | DNA hypermethylation(GO:0044026) hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.3 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.5 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.3 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.2 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.2 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.0 | 0.2 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.2 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.3 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.4 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.1 | GO:1904346 | positive regulation of growth rate(GO:0040010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.0 | 0.1 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.2 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.0 | 0.1 | GO:1904875 | regulation of DNA ligase activity(GO:1904875) |
| 0.0 | 0.1 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.2 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:2000097 | regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.2 | GO:0045337 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.2 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:0030856 | regulation of epithelial cell differentiation(GO:0030856) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.3 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.1 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 0.3 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.3 | GO:0071600 | otic vesicle morphogenesis(GO:0071600) |
| 0.0 | 0.1 | GO:0002778 | antimicrobial peptide production(GO:0002775) antibacterial peptide production(GO:0002778) |
| 0.0 | 0.2 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.3 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.3 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 1.1 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.1 | GO:0072301 | metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) negative regulation of metanephric glomerulus development(GO:0072299) regulation of metanephric glomerular mesangial cell proliferation(GO:0072301) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.1 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.3 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.1 | GO:0086097 | renin-angiotensin regulation of aldosterone production(GO:0002018) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.3 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:2000426 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0070627 | ferrous iron import(GO:0070627) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.1 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.2 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.1 | GO:0048633 | somite specification(GO:0001757) negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) positive regulation of skeletal muscle tissue growth(GO:0048633) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.7 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:1902824 | cleavage furrow ingression(GO:0036090) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.1 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.0 | GO:0046823 | negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein import(GO:1904590) |
| 0.0 | 0.1 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.5 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.3 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.3 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.2 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.1 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.0 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.0 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 0.2 | GO:0071888 | macrophage apoptotic process(GO:0071888) |
| 0.0 | 0.1 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.0 | 0.0 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.1 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.8 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.0 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.0 | 0.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.2 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 0.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.5 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.1 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.5 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.0 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.8 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.6 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.7 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.2 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.1 | 0.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.2 | GO:0004641 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.1 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 1.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) azole transporter activity(GO:0045118) |
| 0.0 | 0.2 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.1 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.0 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.1 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.1 | REACTOME PI3K EVENTS IN ERBB2 SIGNALING | Genes involved in PI3K events in ERBB2 signaling |
| 0.0 | 0.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |