Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
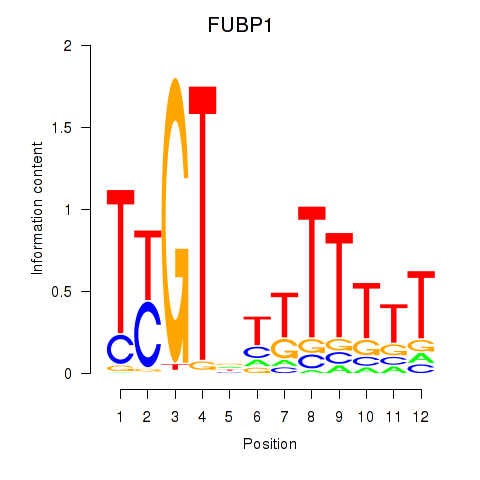
Results for FUBP1
Z-value: 0.53
Transcription factors associated with FUBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FUBP1
|
ENSG00000162613.12 | far upstream element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FUBP1 | hg19_v2_chr1_-_78444776_78444800 | 0.92 | 7.6e-02 | Click! |
Activity profile of FUBP1 motif
Sorted Z-values of FUBP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_61654271 | 0.24 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr2_-_27435390 | 0.22 |
ENST00000428518.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr7_-_75401513 | 0.22 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr2_-_27435634 | 0.21 |
ENST00000430186.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr10_+_17686193 | 0.20 |
ENST00000377500.1
|
STAM
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr11_-_46141338 | 0.19 |
ENST00000529782.1
ENST00000532010.1 ENST00000525438.1 ENST00000533757.1 ENST00000527782.1 |
PHF21A
|
PHD finger protein 21A |
| chr10_+_114710516 | 0.18 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr5_+_72509751 | 0.18 |
ENST00000515556.1
ENST00000513379.1 ENST00000427584.2 |
RP11-60A8.1
|
RP11-60A8.1 |
| chr10_+_17686221 | 0.18 |
ENST00000540523.1
|
STAM
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr1_-_161207986 | 0.18 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr4_+_89444961 | 0.18 |
ENST00000513325.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr12_+_57810198 | 0.17 |
ENST00000598001.1
|
AC126614.1
|
HCG1818482; Uncharacterized protein |
| chr7_-_83824449 | 0.17 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chrX_-_73513353 | 0.16 |
ENST00000430772.1
ENST00000423992.2 ENST00000602812.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr8_+_40018977 | 0.16 |
ENST00000520487.1
|
RP11-470M17.2
|
RP11-470M17.2 |
| chr11_+_110001723 | 0.15 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr2_+_162087577 | 0.15 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr21_+_17791648 | 0.15 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr3_-_71632894 | 0.14 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr9_-_16705069 | 0.14 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr6_-_56716686 | 0.13 |
ENST00000520645.1
|
DST
|
dystonin |
| chr9_+_128510454 | 0.13 |
ENST00000491787.3
ENST00000447726.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr9_+_116173000 | 0.13 |
ENST00000288462.4
|
C9orf43
|
chromosome 9 open reading frame 43 |
| chr17_+_67498538 | 0.13 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr6_-_111888474 | 0.13 |
ENST00000368735.1
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr1_-_154178803 | 0.13 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr3_-_167452262 | 0.11 |
ENST00000487947.2
|
PDCD10
|
programmed cell death 10 |
| chr10_-_70092671 | 0.11 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr6_-_56507586 | 0.11 |
ENST00000439203.1
ENST00000518935.1 ENST00000446842.2 ENST00000370765.6 ENST00000244364.6 |
DST
|
dystonin |
| chr2_-_208030295 | 0.11 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr3_-_112127981 | 0.10 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr16_-_3350614 | 0.10 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr2_-_65593784 | 0.10 |
ENST00000443619.2
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr1_+_45140360 | 0.10 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr1_-_244006528 | 0.09 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr2_-_152118276 | 0.09 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr3_-_167452298 | 0.09 |
ENST00000475915.2
ENST00000462725.2 ENST00000461494.1 |
PDCD10
|
programmed cell death 10 |
| chr12_+_57522692 | 0.09 |
ENST00000554174.1
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr10_+_32856764 | 0.09 |
ENST00000375030.2
ENST00000375028.3 |
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr10_+_79793518 | 0.08 |
ENST00000440692.1
ENST00000435275.1 ENST00000372360.3 ENST00000360830.4 |
RPS24
|
ribosomal protein S24 |
| chr14_-_23762777 | 0.08 |
ENST00000431326.2
|
HOMEZ
|
homeobox and leucine zipper encoding |
| chr14_-_21852119 | 0.08 |
ENST00000555943.1
|
SUPT16H
|
suppressor of Ty 16 homolog (S. cerevisiae) |
| chr12_+_14572070 | 0.08 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr5_-_1882858 | 0.08 |
ENST00000511126.1
ENST00000231357.2 |
IRX4
|
iroquois homeobox 4 |
| chr20_+_9049303 | 0.08 |
ENST00000407043.2
ENST00000441846.1 |
PLCB4
|
phospholipase C, beta 4 |
| chr9_+_96846740 | 0.08 |
ENST00000288976.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr17_-_42298201 | 0.07 |
ENST00000527034.1
|
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr15_-_59500973 | 0.07 |
ENST00000560749.1
|
MYO1E
|
myosin IE |
| chr13_+_44596471 | 0.07 |
ENST00000592085.1
ENST00000591799.1 |
LINC00284
|
long intergenic non-protein coding RNA 284 |
| chr6_-_131949305 | 0.07 |
ENST00000368053.4
ENST00000354577.4 ENST00000403834.3 ENST00000540546.1 ENST00000368068.3 ENST00000368060.3 |
MED23
|
mediator complex subunit 23 |
| chr1_-_182360498 | 0.07 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr10_-_65028817 | 0.07 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr18_-_60986613 | 0.07 |
ENST00000444484.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr15_+_38276910 | 0.07 |
ENST00000558081.2
|
RP11-1008C21.1
|
RP11-1008C21.1 |
| chr1_+_45140400 | 0.06 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr1_-_161208013 | 0.06 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_-_182360918 | 0.06 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr5_+_140027355 | 0.06 |
ENST00000417647.2
ENST00000507593.1 ENST00000508301.1 |
IK
|
IK cytokine, down-regulator of HLA II |
| chr4_+_170581213 | 0.06 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr14_-_94547541 | 0.06 |
ENST00000544005.1
ENST00000555054.1 ENST00000330836.5 |
DDX24
|
DEAD (Asp-Glu-Ala-Asp) box helicase 24 |
| chr10_+_17686124 | 0.06 |
ENST00000377524.3
|
STAM
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr4_-_25865159 | 0.06 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr15_-_89764929 | 0.06 |
ENST00000268125.5
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr4_-_2420335 | 0.06 |
ENST00000503000.1
|
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chr1_+_171107241 | 0.06 |
ENST00000236166.3
|
FMO6P
|
flavin containing monooxygenase 6 pseudogene |
| chr1_-_150738261 | 0.06 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr4_-_99850243 | 0.06 |
ENST00000280892.6
ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr1_-_198906528 | 0.06 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr17_-_42298331 | 0.05 |
ENST00000343638.5
|
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr1_-_27701307 | 0.05 |
ENST00000270879.4
ENST00000354982.2 |
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 |
| chr2_-_216300784 | 0.05 |
ENST00000421182.1
ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1
|
fibronectin 1 |
| chr5_+_145316120 | 0.05 |
ENST00000359120.4
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr20_+_9049742 | 0.05 |
ENST00000437503.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr19_+_48969094 | 0.05 |
ENST00000595676.1
|
CTC-273B12.7
|
Uncharacterized protein |
| chr12_+_57522439 | 0.05 |
ENST00000338962.4
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr5_+_180682720 | 0.05 |
ENST00000599439.1
|
AC008443.1
|
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr4_-_48136217 | 0.05 |
ENST00000264316.4
|
TXK
|
TXK tyrosine kinase |
| chr12_+_12870055 | 0.05 |
ENST00000228872.4
|
CDKN1B
|
cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
| chr11_-_46142615 | 0.04 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr7_-_74267836 | 0.04 |
ENST00000361071.5
ENST00000453619.2 ENST00000417115.2 ENST00000405086.2 |
GTF2IRD2
|
GTF2I repeat domain containing 2 |
| chr1_-_248005254 | 0.04 |
ENST00000355784.2
|
OR11L1
|
olfactory receptor, family 11, subfamily L, member 1 |
| chr14_+_89060739 | 0.04 |
ENST00000318308.6
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr20_+_9049682 | 0.04 |
ENST00000334005.3
ENST00000378473.3 |
PLCB4
|
phospholipase C, beta 4 |
| chr9_-_123812542 | 0.04 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr1_+_43855545 | 0.04 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr12_+_57522258 | 0.04 |
ENST00000553277.1
ENST00000243077.3 |
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr9_+_4662282 | 0.04 |
ENST00000381883.2
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2 |
| chrX_+_95939638 | 0.04 |
ENST00000373061.3
ENST00000373054.4 ENST00000355827.4 |
DIAPH2
|
diaphanous-related formin 2 |
| chr15_+_49462397 | 0.04 |
ENST00000396509.2
|
GALK2
|
galactokinase 2 |
| chr1_+_202317815 | 0.04 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr10_-_32667660 | 0.04 |
ENST00000375110.2
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila) |
| chr4_-_101439242 | 0.04 |
ENST00000296420.4
|
EMCN
|
endomucin |
| chr14_-_38036271 | 0.04 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr12_-_26986076 | 0.04 |
ENST00000381340.3
|
ITPR2
|
inositol 1,4,5-trisphosphate receptor, type 2 |
| chr7_-_15601595 | 0.04 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr13_-_26795840 | 0.04 |
ENST00000381570.3
ENST00000399762.2 ENST00000346166.3 |
RNF6
|
ring finger protein (C3H2C3 type) 6 |
| chr15_+_52155001 | 0.04 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr1_+_205682497 | 0.04 |
ENST00000598338.1
|
AC119673.1
|
AC119673.1 |
| chr16_+_24741013 | 0.04 |
ENST00000315183.7
ENST00000395799.3 |
TNRC6A
|
trinucleotide repeat containing 6A |
| chr10_+_98592674 | 0.04 |
ENST00000356016.3
ENST00000371097.4 |
LCOR
|
ligand dependent nuclear receptor corepressor |
| chr14_+_76072096 | 0.04 |
ENST00000555058.1
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr3_-_170303845 | 0.04 |
ENST00000231706.5
|
SLC7A14
|
solute carrier family 7, member 14 |
| chr15_+_49462434 | 0.04 |
ENST00000558145.1
ENST00000543495.1 ENST00000544523.1 ENST00000560138.1 |
GALK2
|
galactokinase 2 |
| chr7_+_129710350 | 0.04 |
ENST00000335420.5
ENST00000463413.1 |
KLHDC10
|
kelch domain containing 10 |
| chr11_-_33891362 | 0.03 |
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr1_-_245134273 | 0.03 |
ENST00000607453.1
|
RP11-156E8.1
|
Uncharacterized protein |
| chr22_-_29137771 | 0.03 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr3_-_71114066 | 0.03 |
ENST00000485326.2
|
FOXP1
|
forkhead box P1 |
| chrX_-_73513307 | 0.03 |
ENST00000602420.1
|
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr2_-_183291741 | 0.03 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr5_+_125706998 | 0.03 |
ENST00000506445.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr7_-_144435985 | 0.03 |
ENST00000549981.1
|
TPK1
|
thiamin pyrophosphokinase 1 |
| chr9_-_113761720 | 0.03 |
ENST00000541779.1
ENST00000374430.2 |
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr1_+_209602771 | 0.03 |
ENST00000440276.1
|
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr7_-_83824169 | 0.03 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_+_7836398 | 0.03 |
ENST00000565740.1
|
CNTROB
|
centrobin, centrosomal BRCA2 interacting protein |
| chr21_+_17792672 | 0.03 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr10_+_101542462 | 0.03 |
ENST00000370449.4
ENST00000370434.1 |
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr12_+_56324756 | 0.03 |
ENST00000331886.5
ENST00000555090.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr5_-_34919094 | 0.03 |
ENST00000341754.4
|
RAD1
|
RAD1 homolog (S. pombe) |
| chrX_+_95939711 | 0.03 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr11_+_120207787 | 0.03 |
ENST00000397843.2
ENST00000356641.3 |
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr12_-_57522813 | 0.03 |
ENST00000556155.1
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr1_+_78383813 | 0.03 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr5_-_159846399 | 0.03 |
ENST00000297151.4
|
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr3_-_149510553 | 0.03 |
ENST00000462519.2
ENST00000446160.1 ENST00000383050.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chrX_+_119737806 | 0.02 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr11_-_46142505 | 0.02 |
ENST00000524497.1
ENST00000418153.2 |
PHF21A
|
PHD finger protein 21A |
| chr2_+_201980827 | 0.02 |
ENST00000309955.3
ENST00000443227.1 ENST00000341222.6 ENST00000355558.4 ENST00000340870.5 ENST00000341582.6 |
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr19_+_10765003 | 0.02 |
ENST00000407004.3
ENST00000589998.1 ENST00000589600.1 |
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr8_+_94929969 | 0.02 |
ENST00000517764.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr7_+_35756092 | 0.02 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chr2_+_202122826 | 0.02 |
ENST00000413726.1
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr10_+_51187938 | 0.02 |
ENST00000311663.5
|
FAM21D
|
family with sequence similarity 21, member D |
| chr7_+_137761199 | 0.02 |
ENST00000411726.2
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr4_-_89152474 | 0.02 |
ENST00000515655.1
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr21_+_45209394 | 0.02 |
ENST00000497547.1
|
RRP1
|
ribosomal RNA processing 1 |
| chr1_-_75139397 | 0.02 |
ENST00000326665.5
|
C1orf173
|
chromosome 1 open reading frame 173 |
| chr19_+_13134772 | 0.02 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr11_+_34073195 | 0.02 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr3_-_182881595 | 0.02 |
ENST00000476015.1
|
LAMP3
|
lysosomal-associated membrane protein 3 |
| chr12_+_10365404 | 0.02 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr2_+_113033164 | 0.02 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr3_+_57875738 | 0.02 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chr14_+_65878650 | 0.02 |
ENST00000555559.1
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr3_-_112693759 | 0.02 |
ENST00000440122.2
ENST00000490004.1 |
CD200R1
|
CD200 receptor 1 |
| chr9_-_119162885 | 0.02 |
ENST00000445861.2
|
PAPPA-AS1
|
PAPPA antisense RNA 1 |
| chr11_-_47521309 | 0.02 |
ENST00000535982.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr19_-_55453077 | 0.02 |
ENST00000328092.5
ENST00000590030.1 |
NLRP7
|
NLR family, pyrin domain containing 7 |
| chr1_+_109102652 | 0.02 |
ENST00000370035.3
ENST00000405454.1 |
FAM102B
|
family with sequence similarity 102, member B |
| chr3_+_19988566 | 0.02 |
ENST00000273047.4
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chrX_+_22056165 | 0.02 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr7_-_87849340 | 0.01 |
ENST00000419179.1
ENST00000265729.2 |
SRI
|
sorcin |
| chr2_+_202122703 | 0.01 |
ENST00000447616.1
ENST00000358485.4 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr12_+_48513570 | 0.01 |
ENST00000551804.1
|
PFKM
|
phosphofructokinase, muscle |
| chr7_+_74508372 | 0.01 |
ENST00000356115.5
ENST00000430511.2 ENST00000312575.7 |
GTF2IRD2B
|
GTF2I repeat domain containing 2B |
| chr19_-_6057282 | 0.01 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr7_+_116166331 | 0.01 |
ENST00000393468.1
ENST00000393467.1 |
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr19_+_10765614 | 0.01 |
ENST00000589283.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr18_+_7754957 | 0.01 |
ENST00000400053.4
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr7_-_22233442 | 0.01 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr12_+_27863706 | 0.01 |
ENST00000081029.3
ENST00000538315.1 ENST00000542791.1 |
MRPS35
|
mitochondrial ribosomal protein S35 |
| chr1_+_65613513 | 0.01 |
ENST00000395334.2
|
AK4
|
adenylate kinase 4 |
| chr1_+_110158726 | 0.01 |
ENST00000531734.1
|
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr3_-_187454281 | 0.01 |
ENST00000232014.4
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr1_+_218519577 | 0.01 |
ENST00000366930.4
ENST00000366929.4 |
TGFB2
|
transforming growth factor, beta 2 |
| chr12_-_110888103 | 0.01 |
ENST00000426440.1
ENST00000228825.7 |
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr12_+_121088291 | 0.01 |
ENST00000351200.2
|
CABP1
|
calcium binding protein 1 |
| chr20_-_45981138 | 0.01 |
ENST00000446994.2
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr15_+_49170083 | 0.01 |
ENST00000530028.2
|
EID1
|
EP300 interacting inhibitor of differentiation 1 |
| chr6_+_119215308 | 0.01 |
ENST00000229595.5
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr14_+_58797974 | 0.01 |
ENST00000417477.2
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr1_+_89149905 | 0.01 |
ENST00000316005.7
ENST00000370521.3 ENST00000370505.3 |
PKN2
|
protein kinase N2 |
| chr7_-_22234381 | 0.01 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr2_-_175202151 | 0.01 |
ENST00000595354.1
|
AC018470.1
|
Uncharacterized protein FLJ46347 |
| chr15_-_88799948 | 0.01 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr13_+_49684445 | 0.01 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr5_+_156696362 | 0.01 |
ENST00000377576.3
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr10_-_56561022 | 0.01 |
ENST00000373965.2
ENST00000414778.1 ENST00000395438.1 ENST00000409834.1 ENST00000395445.1 ENST00000395446.1 ENST00000395442.1 ENST00000395440.1 ENST00000395432.2 ENST00000361849.3 ENST00000395433.1 ENST00000320301.6 ENST00000395430.1 ENST00000437009.1 |
PCDH15
|
protocadherin-related 15 |
| chr10_-_91102410 | 0.01 |
ENST00000282673.4
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase |
| chr5_+_156712372 | 0.00 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr12_+_56324933 | 0.00 |
ENST00000549629.1
ENST00000555218.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr7_-_127032114 | 0.00 |
ENST00000436992.1
|
ZNF800
|
zinc finger protein 800 |
| chr16_+_28565230 | 0.00 |
ENST00000317058.3
|
CCDC101
|
coiled-coil domain containing 101 |
| chr4_-_168155730 | 0.00 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr3_-_196242233 | 0.00 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr1_-_155271213 | 0.00 |
ENST00000342741.4
|
PKLR
|
pyruvate kinase, liver and RBC |
| chr1_+_228870824 | 0.00 |
ENST00000366691.3
|
RHOU
|
ras homolog family member U |
| chr21_-_38445297 | 0.00 |
ENST00000430792.1
ENST00000399103.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr4_-_152149033 | 0.00 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr7_-_124405681 | 0.00 |
ENST00000303921.2
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like) |
| chr1_+_181452678 | 0.00 |
ENST00000367570.1
ENST00000526775.1 ENST00000357570.5 ENST00000358338.5 ENST00000367567.4 |
CACNA1E
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr18_-_10855434 | 0.00 |
ENST00000579112.1
|
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr7_+_141811539 | 0.00 |
ENST00000550469.2
ENST00000477922.3 |
RP11-1220K2.2
|
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr21_+_17791838 | 0.00 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr16_+_81070792 | 0.00 |
ENST00000564241.1
ENST00000565237.1 |
ATMIN
|
ATM interactor |
| chr13_+_49551020 | 0.00 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr18_-_60985914 | 0.00 |
ENST00000589955.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FUBP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.0 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.4 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.1 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.2 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.2 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.1 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.0 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.0 | GO:0005579 | membrane attack complex(GO:0005579) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.2 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.1 | 0.4 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.2 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |