Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
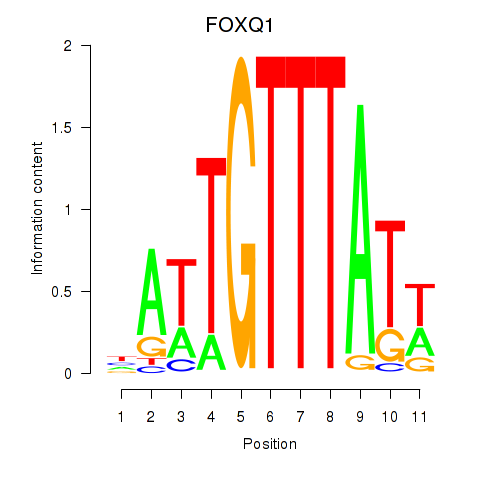
Results for FOXQ1
Z-value: 0.70
Transcription factors associated with FOXQ1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXQ1
|
ENSG00000164379.4 | forkhead box Q1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXQ1 | hg19_v2_chr6_+_1312675_1312701 | -0.75 | 2.5e-01 | Click! |
Activity profile of FOXQ1 motif
Sorted Z-values of FOXQ1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_78505581 | 0.53 |
ENST00000376767.3
ENST00000376752.4 |
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr14_+_52456327 | 0.49 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr1_+_104159999 | 0.45 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr19_+_7580103 | 0.40 |
ENST00000596712.1
|
ZNF358
|
zinc finger protein 358 |
| chr3_+_193853927 | 0.34 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr11_+_71544246 | 0.30 |
ENST00000328698.1
|
DEFB108B
|
defensin, beta 108B |
| chr8_+_52730143 | 0.30 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr17_-_46690839 | 0.29 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr15_+_63188009 | 0.28 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr10_-_36813162 | 0.28 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr15_+_49715293 | 0.27 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr4_-_111120334 | 0.23 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr14_+_52456193 | 0.22 |
ENST00000261700.3
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr1_+_28099700 | 0.22 |
ENST00000440806.2
|
STX12
|
syntaxin 12 |
| chr2_-_4703793 | 0.22 |
ENST00000421212.1
ENST00000412134.1 |
AC022311.1
|
AC022311.1 |
| chr10_-_10504285 | 0.22 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr16_-_69418553 | 0.21 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr4_+_165675197 | 0.21 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr15_+_35270552 | 0.21 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr4_+_124571409 | 0.21 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr15_+_80351977 | 0.20 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr18_+_19668021 | 0.20 |
ENST00000579830.1
|
RP11-595B24.2
|
Uncharacterized protein |
| chr1_+_28099683 | 0.20 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chr3_-_57233966 | 0.19 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr9_-_98268883 | 0.19 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chrX_-_139866723 | 0.19 |
ENST00000370532.2
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa |
| chr5_+_81601166 | 0.19 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr6_+_123038689 | 0.18 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr3_-_156840776 | 0.18 |
ENST00000471357.1
|
LINC00880
|
long intergenic non-protein coding RNA 880 |
| chr2_+_191208791 | 0.18 |
ENST00000423767.1
ENST00000451089.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr12_-_102133191 | 0.18 |
ENST00000392924.1
ENST00000266743.2 ENST00000392927.3 |
SYCP3
|
synaptonemal complex protein 3 |
| chr14_+_56584414 | 0.18 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr12_-_110511424 | 0.18 |
ENST00000548191.1
|
C12orf76
|
chromosome 12 open reading frame 76 |
| chr15_+_49715449 | 0.18 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr15_+_31658349 | 0.18 |
ENST00000558844.1
|
KLF13
|
Kruppel-like factor 13 |
| chr7_-_64023441 | 0.18 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr8_+_142264664 | 0.18 |
ENST00000518520.1
|
RP11-10J21.3
|
Uncharacterized protein |
| chr5_+_136070614 | 0.17 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr10_+_5454505 | 0.17 |
ENST00000355029.4
|
NET1
|
neuroepithelial cell transforming 1 |
| chr1_+_162602244 | 0.17 |
ENST00000367922.3
ENST00000367921.3 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr22_-_39637135 | 0.17 |
ENST00000440375.1
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr12_-_102591604 | 0.17 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr4_+_146403912 | 0.16 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr12_-_76478386 | 0.16 |
ENST00000535020.2
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr5_-_159846066 | 0.16 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr4_-_74853897 | 0.16 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr18_+_11752783 | 0.16 |
ENST00000585642.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr1_+_145525015 | 0.16 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr6_+_116575329 | 0.15 |
ENST00000430252.2
ENST00000540275.1 ENST00000448740.2 |
DSE
RP3-486I3.7
|
dermatan sulfate epimerase RP3-486I3.7 |
| chr12_-_76478417 | 0.15 |
ENST00000552342.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr5_-_60458179 | 0.15 |
ENST00000507416.1
ENST00000339020.3 |
SMIM15
|
small integral membrane protein 15 |
| chr1_+_120839412 | 0.15 |
ENST00000355228.4
|
FAM72B
|
family with sequence similarity 72, member B |
| chr4_-_70626430 | 0.15 |
ENST00000310613.3
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr6_-_31648150 | 0.15 |
ENST00000375858.3
ENST00000383237.4 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr3_-_96337000 | 0.15 |
ENST00000600213.2
|
MTRNR2L12
|
MT-RNR2-like 12 (pseudogene) |
| chr4_-_80994210 | 0.15 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr21_+_17443521 | 0.14 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr14_-_92247032 | 0.14 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr15_-_76352069 | 0.14 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr3_+_159570722 | 0.14 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr17_+_20978854 | 0.14 |
ENST00000456235.1
|
AC087393.1
|
AC087393.1 |
| chr6_-_138833630 | 0.13 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr19_+_37178482 | 0.13 |
ENST00000536254.2
|
ZNF567
|
zinc finger protein 567 |
| chr4_-_103266626 | 0.13 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr6_+_24777040 | 0.13 |
ENST00000378059.3
|
GMNN
|
geminin, DNA replication inhibitor |
| chr16_-_15474904 | 0.13 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr21_+_25801041 | 0.13 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr12_-_76478446 | 0.13 |
ENST00000393263.3
ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr10_+_35484793 | 0.13 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr7_-_64023410 | 0.13 |
ENST00000447137.2
|
ZNF680
|
zinc finger protein 680 |
| chr4_-_111120132 | 0.12 |
ENST00000506625.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr2_+_95873257 | 0.12 |
ENST00000425953.1
|
AC092835.2
|
AC092835.2 |
| chr1_+_231473990 | 0.12 |
ENST00000008440.9
|
SPRTN
|
SprT-like N-terminal domain |
| chr10_+_70847852 | 0.12 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr4_-_111119804 | 0.12 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr18_+_3252265 | 0.12 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr10_-_74283694 | 0.12 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr20_+_30327063 | 0.11 |
ENST00000300403.6
ENST00000340513.4 |
TPX2
|
TPX2, microtubule-associated |
| chr11_+_112047087 | 0.11 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr19_-_14992264 | 0.11 |
ENST00000327462.2
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17 |
| chr12_-_76461249 | 0.11 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr1_-_145826450 | 0.11 |
ENST00000462900.2
|
GPR89A
|
G protein-coupled receptor 89A |
| chr15_+_80351910 | 0.11 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr6_+_22221010 | 0.11 |
ENST00000567753.1
|
RP11-524C21.2
|
RP11-524C21.2 |
| chr4_-_52883786 | 0.11 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr15_-_59041768 | 0.11 |
ENST00000402627.1
ENST00000396140.2 ENST00000559053.1 ENST00000561288.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr16_+_28763108 | 0.11 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr22_-_39636914 | 0.11 |
ENST00000381551.4
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr1_+_239882842 | 0.11 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr1_-_33430286 | 0.11 |
ENST00000373456.7
ENST00000356990.5 ENST00000235150.4 |
RNF19B
|
ring finger protein 19B |
| chr22_+_23029188 | 0.11 |
ENST00000390305.2
|
IGLV3-25
|
immunoglobulin lambda variable 3-25 |
| chr3_-_128902759 | 0.11 |
ENST00000422453.2
ENST00000504813.1 ENST00000512338.1 |
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein |
| chr5_+_154173697 | 0.11 |
ENST00000518742.1
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr14_-_35591156 | 0.11 |
ENST00000554361.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr8_+_110551925 | 0.10 |
ENST00000395785.2
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr7_+_115862858 | 0.10 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr19_-_39402798 | 0.10 |
ENST00000571838.1
|
CTC-360G5.1
|
coiled-coil glutamate-rich protein 2 |
| chr18_+_61143994 | 0.10 |
ENST00000382771.4
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr12_-_96389702 | 0.10 |
ENST00000552509.1
|
HAL
|
histidine ammonia-lyase |
| chr10_+_5488564 | 0.10 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr11_-_62457371 | 0.10 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr15_+_41549105 | 0.10 |
ENST00000560965.1
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr3_-_128902729 | 0.10 |
ENST00000451728.2
ENST00000446936.2 ENST00000502976.1 ENST00000500450.2 ENST00000441626.2 |
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein |
| chr18_-_57027194 | 0.10 |
ENST00000251047.5
|
LMAN1
|
lectin, mannose-binding, 1 |
| chr19_-_48823332 | 0.10 |
ENST00000315396.7
|
CCDC114
|
coiled-coil domain containing 114 |
| chr8_+_109455845 | 0.10 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr3_+_171561127 | 0.10 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr1_+_145524891 | 0.10 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr12_+_56435637 | 0.10 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chr2_+_74120094 | 0.10 |
ENST00000409731.3
ENST00000345517.3 ENST00000409918.1 ENST00000442912.1 ENST00000409624.1 |
ACTG2
|
actin, gamma 2, smooth muscle, enteric |
| chr10_+_86184676 | 0.10 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr12_-_8803128 | 0.10 |
ENST00000543467.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr4_-_120225600 | 0.10 |
ENST00000399075.4
|
C4orf3
|
chromosome 4 open reading frame 3 |
| chr1_-_151148492 | 0.10 |
ENST00000295314.4
|
TMOD4
|
tropomodulin 4 (muscle) |
| chr4_-_119274121 | 0.10 |
ENST00000296498.3
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr2_-_151344172 | 0.10 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr1_-_153599732 | 0.10 |
ENST00000392623.1
|
S100A13
|
S100 calcium binding protein A13 |
| chr18_-_52626622 | 0.10 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr3_-_167191814 | 0.10 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr6_+_37321823 | 0.10 |
ENST00000487950.1
ENST00000469731.1 |
RNF8
|
ring finger protein 8, E3 ubiquitin protein ligase |
| chr10_-_104913367 | 0.09 |
ENST00000423468.2
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr4_-_147442982 | 0.09 |
ENST00000511374.1
ENST00000264986.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr2_+_190306159 | 0.09 |
ENST00000314761.4
|
WDR75
|
WD repeat domain 75 |
| chr15_+_50474385 | 0.09 |
ENST00000267842.5
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2 |
| chr3_+_178866199 | 0.09 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr14_+_24867992 | 0.09 |
ENST00000382554.3
|
NYNRIN
|
NYN domain and retroviral integrase containing |
| chr17_+_45771420 | 0.09 |
ENST00000578982.1
|
TBKBP1
|
TBK1 binding protein 1 |
| chr4_-_147443043 | 0.09 |
ENST00000394059.4
ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr4_+_75174204 | 0.09 |
ENST00000332112.4
ENST00000514968.1 ENST00000503098.1 ENST00000502358.1 ENST00000509145.1 ENST00000505212.1 |
EPGN
|
epithelial mitogen |
| chr21_-_16254231 | 0.09 |
ENST00000412426.1
ENST00000418954.1 |
AF127936.7
|
AF127936.7 |
| chr1_+_221054411 | 0.09 |
ENST00000427693.1
|
HLX
|
H2.0-like homeobox |
| chr14_+_56078695 | 0.09 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr3_+_182983090 | 0.09 |
ENST00000465010.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr16_-_25122735 | 0.09 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr2_-_188419200 | 0.09 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr11_+_108535752 | 0.09 |
ENST00000322536.3
|
DDX10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
| chr2_-_218843623 | 0.09 |
ENST00000413280.1
|
TNS1
|
tensin 1 |
| chr10_-_91295304 | 0.09 |
ENST00000341233.4
ENST00000371790.4 |
SLC16A12
|
solute carrier family 16, member 12 |
| chr15_-_55488817 | 0.09 |
ENST00000569386.1
|
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr11_-_64163297 | 0.09 |
ENST00000457725.1
|
AP003774.6
|
AP003774.6 |
| chr2_-_225434538 | 0.09 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr20_+_34802295 | 0.08 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr1_-_150669604 | 0.08 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr2_+_66918558 | 0.08 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr12_+_27396901 | 0.08 |
ENST00000541191.1
ENST00000389032.3 |
STK38L
|
serine/threonine kinase 38 like |
| chr12_+_109490370 | 0.08 |
ENST00000257548.5
ENST00000536723.1 ENST00000536393.1 |
USP30
|
ubiquitin specific peptidase 30 |
| chr9_-_88897426 | 0.08 |
ENST00000375991.4
ENST00000326094.4 |
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr1_+_26605618 | 0.08 |
ENST00000270792.5
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr4_+_86749045 | 0.08 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr3_+_101280677 | 0.08 |
ENST00000309922.6
ENST00000495642.1 |
TRMT10C
|
tRNA methyltransferase 10 homolog C (S. cerevisiae) |
| chr4_+_95174445 | 0.08 |
ENST00000509418.1
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr1_+_145883868 | 0.08 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr5_+_64920543 | 0.08 |
ENST00000399438.3
ENST00000510585.2 |
TRAPPC13
CTC-534A2.2
|
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chrX_+_108780347 | 0.08 |
ENST00000372103.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr3_-_145940126 | 0.08 |
ENST00000498625.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr14_+_77843459 | 0.08 |
ENST00000216471.4
|
SAMD15
|
sterile alpha motif domain containing 15 |
| chr3_+_157154578 | 0.08 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr4_-_147442817 | 0.08 |
ENST00000507030.1
|
SLC10A7
|
solute carrier family 10, member 7 |
| chr2_+_163175394 | 0.08 |
ENST00000446271.1
ENST00000429691.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr12_-_49582593 | 0.08 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr1_-_95391315 | 0.08 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr11_-_82997420 | 0.08 |
ENST00000455220.2
ENST00000529689.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr21_+_30968360 | 0.08 |
ENST00000333765.4
|
GRIK1-AS2
|
GRIK1 antisense RNA 2 |
| chr1_-_115259337 | 0.08 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr4_-_80994619 | 0.08 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr2_-_192711968 | 0.08 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr5_+_118690466 | 0.08 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr21_-_35899113 | 0.08 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr5_+_118812294 | 0.08 |
ENST00000509514.1
|
HSD17B4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr11_-_68611721 | 0.08 |
ENST00000561996.1
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver) |
| chr8_+_79428539 | 0.08 |
ENST00000352966.5
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr1_-_100643765 | 0.08 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr5_+_218356 | 0.08 |
ENST00000264932.6
ENST00000504309.1 ENST00000510361.1 |
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr1_+_204839959 | 0.08 |
ENST00000404076.1
|
NFASC
|
neurofascin |
| chrM_+_5824 | 0.07 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr10_+_5090940 | 0.07 |
ENST00000602997.1
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr11_+_45943169 | 0.07 |
ENST00000529052.1
ENST00000531526.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr7_-_140624499 | 0.07 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr19_-_50528392 | 0.07 |
ENST00000600137.1
ENST00000597215.1 |
VRK3
|
vaccinia related kinase 3 |
| chr5_+_42756903 | 0.07 |
ENST00000361970.5
ENST00000388827.4 |
CCDC152
|
coiled-coil domain containing 152 |
| chr18_+_21032781 | 0.07 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr7_+_142985308 | 0.07 |
ENST00000310447.5
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr7_+_1272522 | 0.07 |
ENST00000316333.8
|
UNCX
|
UNC homeobox |
| chr4_-_21950356 | 0.07 |
ENST00000447367.2
ENST00000382152.2 |
KCNIP4
|
Kv channel interacting protein 4 |
| chr1_+_236686717 | 0.07 |
ENST00000341872.6
ENST00000450372.2 |
LGALS8
|
lectin, galactoside-binding, soluble, 8 |
| chr18_+_7754957 | 0.07 |
ENST00000400053.4
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr8_+_94767072 | 0.07 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr1_-_227505289 | 0.07 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr8_-_82644562 | 0.07 |
ENST00000520604.1
ENST00000521742.1 ENST00000520635.1 |
ZFAND1
|
zinc finger, AN1-type domain 1 |
| chr22_+_24828087 | 0.07 |
ENST00000472248.1
ENST00000436735.1 |
ADORA2A
|
adenosine A2a receptor |
| chrX_+_37208521 | 0.07 |
ENST00000378628.4
|
PRRG1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr8_-_124037890 | 0.07 |
ENST00000519018.1
ENST00000523036.1 |
DERL1
|
derlin 1 |
| chr16_-_2827128 | 0.07 |
ENST00000494946.2
ENST00000409477.1 ENST00000572954.1 ENST00000262306.7 ENST00000409906.4 |
TCEB2
|
transcription elongation factor B (SIII), polypeptide 2 (18kDa, elongin B) |
| chr17_-_36358166 | 0.07 |
ENST00000537432.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr8_+_30013813 | 0.07 |
ENST00000221114.3
|
DCTN6
|
dynactin 6 |
| chr14_+_102276209 | 0.07 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr2_+_201450591 | 0.07 |
ENST00000374700.2
|
AOX1
|
aldehyde oxidase 1 |
| chr2_-_157198860 | 0.07 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr5_-_218251 | 0.07 |
ENST00000296824.3
|
CCDC127
|
coiled-coil domain containing 127 |
| chr14_+_23563952 | 0.07 |
ENST00000319074.4
|
C14orf119
|
chromosome 14 open reading frame 119 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXQ1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.3 | GO:0060164 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) regulation of timing of neuron differentiation(GO:0060164) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.1 | 0.6 | GO:0001999 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) renin secretion into blood stream(GO:0002001) |
| 0.1 | 0.4 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.3 | GO:0072255 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.1 | 0.2 | GO:1903770 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.3 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.5 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:1900005 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.1 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.1 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.0 | GO:0099553 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.0 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.0 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0070885 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.0 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.0 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.3 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.1 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.0 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.0 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.5 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.2 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.1 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.3 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.1 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0046790 | complement component C1q binding(GO:0001849) virion binding(GO:0046790) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.5 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |