Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
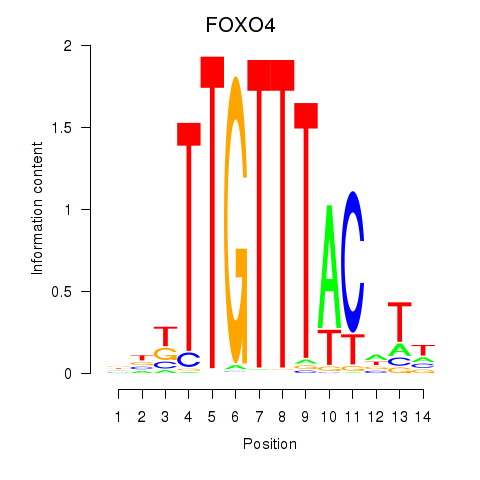
Results for FOXO4
Z-value: 0.45
Transcription factors associated with FOXO4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXO4
|
ENSG00000184481.12 | forkhead box O4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXO4 | hg19_v2_chrX_+_70316005_70316047 | 0.72 | 2.8e-01 | Click! |
Activity profile of FOXO4 motif
Sorted Z-values of FOXO4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_53303123 | 0.28 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr11_+_127140956 | 0.25 |
ENST00000608214.1
|
RP11-480C22.1
|
RP11-480C22.1 |
| chr19_+_859654 | 0.23 |
ENST00000592860.1
|
CFD
|
complement factor D (adipsin) |
| chr17_+_72426891 | 0.18 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr3_-_134092561 | 0.18 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr15_+_75940218 | 0.18 |
ENST00000308527.5
|
SNX33
|
sorting nexin 33 |
| chr11_+_844406 | 0.17 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr16_+_77233294 | 0.17 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr11_+_844067 | 0.17 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr8_-_145754428 | 0.17 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr4_-_140223614 | 0.17 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr14_-_61124977 | 0.16 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr7_+_76101379 | 0.15 |
ENST00000429179.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr3_-_129375556 | 0.14 |
ENST00000510323.1
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr14_-_103987679 | 0.14 |
ENST00000553610.1
|
CKB
|
creatine kinase, brain |
| chr3_+_108015382 | 0.14 |
ENST00000463019.3
ENST00000491820.1 ENST00000467562.1 ENST00000482430.1 ENST00000462629.1 |
HHLA2
|
HERV-H LTR-associating 2 |
| chr17_-_79212825 | 0.13 |
ENST00000374769.2
|
ENTHD2
|
ENTH domain containing 2 |
| chr5_+_137673200 | 0.13 |
ENST00000434981.2
|
FAM53C
|
family with sequence similarity 53, member C |
| chr16_-_4852915 | 0.13 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr6_+_143447322 | 0.13 |
ENST00000458219.1
|
AIG1
|
androgen-induced 1 |
| chr7_+_134832808 | 0.13 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr6_+_151358048 | 0.13 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr19_+_859425 | 0.12 |
ENST00000327726.6
|
CFD
|
complement factor D (adipsin) |
| chr1_+_81106951 | 0.12 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr3_+_112930946 | 0.12 |
ENST00000462425.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr17_-_79212884 | 0.12 |
ENST00000300714.3
|
ENTHD2
|
ENTH domain containing 2 |
| chr11_-_62476965 | 0.12 |
ENST00000405837.1
ENST00000531524.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr8_+_39972170 | 0.12 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr7_+_37723420 | 0.11 |
ENST00000476620.1
|
EPDR1
|
ependymin related 1 |
| chr17_+_72427477 | 0.11 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr1_+_153940713 | 0.11 |
ENST00000368601.1
ENST00000368603.1 ENST00000368600.3 |
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr14_-_99737822 | 0.11 |
ENST00000345514.2
ENST00000443726.2 |
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr17_-_40134339 | 0.11 |
ENST00000587727.1
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr1_+_28844778 | 0.11 |
ENST00000411533.1
|
RCC1
|
regulator of chromosome condensation 1 |
| chr19_+_50380917 | 0.10 |
ENST00000535102.2
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr11_-_62477103 | 0.10 |
ENST00000532818.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr1_-_168106536 | 0.10 |
ENST00000537209.1
ENST00000361697.2 ENST00000546300.1 ENST00000367835.1 |
GPR161
|
G protein-coupled receptor 161 |
| chr2_+_58655461 | 0.10 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr12_+_53491220 | 0.10 |
ENST00000548547.1
ENST00000301464.3 |
IGFBP6
|
insulin-like growth factor binding protein 6 |
| chr11_-_46638720 | 0.10 |
ENST00000326737.3
|
HARBI1
|
harbinger transposase derived 1 |
| chr4_+_40198527 | 0.10 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr19_+_50380682 | 0.10 |
ENST00000221543.5
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr5_+_137673945 | 0.10 |
ENST00000513056.1
ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53, member C |
| chr11_+_34999328 | 0.10 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr1_+_43855560 | 0.10 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr5_+_140529630 | 0.10 |
ENST00000543635.1
|
PCDHB6
|
protocadherin beta 6 |
| chr2_+_66918558 | 0.10 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr2_+_196522032 | 0.10 |
ENST00000418005.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr2_-_206950996 | 0.10 |
ENST00000414320.1
|
INO80D
|
INO80 complex subunit D |
| chr12_-_102591604 | 0.09 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr5_-_125930877 | 0.09 |
ENST00000510111.2
ENST00000509270.1 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr12_+_28605426 | 0.09 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr5_+_176853669 | 0.09 |
ENST00000355472.5
|
GRK6
|
G protein-coupled receptor kinase 6 |
| chr19_-_50380536 | 0.09 |
ENST00000391832.3
ENST00000391834.2 ENST00000344175.5 |
AKT1S1
|
AKT1 substrate 1 (proline-rich) |
| chr7_-_37026108 | 0.09 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr3_+_121289551 | 0.09 |
ENST00000334384.3
|
ARGFX
|
arginine-fifty homeobox |
| chr11_+_33061336 | 0.09 |
ENST00000602733.1
|
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr14_-_99737565 | 0.09 |
ENST00000357195.3
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr1_-_153940097 | 0.09 |
ENST00000413622.1
ENST00000310483.6 |
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr8_+_101349823 | 0.09 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr11_+_77532233 | 0.09 |
ENST00000525409.1
|
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr1_-_47655686 | 0.09 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr17_+_79953310 | 0.08 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr20_-_62582475 | 0.08 |
ENST00000369908.5
|
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr11_-_62476694 | 0.08 |
ENST00000524862.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr17_-_46806540 | 0.08 |
ENST00000290295.7
|
HOXB13
|
homeobox B13 |
| chr11_-_62477041 | 0.08 |
ENST00000433053.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr3_-_139195350 | 0.08 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr11_-_62477313 | 0.08 |
ENST00000464544.1
ENST00000530009.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr1_-_16400086 | 0.08 |
ENST00000375662.4
|
FAM131C
|
family with sequence similarity 131, member C |
| chr9_-_106087316 | 0.08 |
ENST00000411575.1
|
RP11-341A22.2
|
RP11-341A22.2 |
| chr12_-_7596735 | 0.08 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr3_-_52868931 | 0.08 |
ENST00000486659.1
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr17_+_79213039 | 0.08 |
ENST00000431388.2
|
C17orf89
|
chromosome 17 open reading frame 89 |
| chr14_+_23067166 | 0.07 |
ENST00000216327.6
ENST00000542041.1 |
ABHD4
|
abhydrolase domain containing 4 |
| chr12_-_111142729 | 0.07 |
ENST00000546713.1
|
HVCN1
|
hydrogen voltage-gated channel 1 |
| chr14_+_70918874 | 0.07 |
ENST00000603540.1
|
ADAM21
|
ADAM metallopeptidase domain 21 |
| chr6_-_32122106 | 0.07 |
ENST00000428778.1
|
PRRT1
|
proline-rich transmembrane protein 1 |
| chr11_+_46638805 | 0.07 |
ENST00000434074.1
ENST00000312040.4 ENST00000451945.1 |
ATG13
|
autophagy related 13 |
| chr14_+_50234827 | 0.07 |
ENST00000554589.1
ENST00000557247.1 |
KLHDC2
|
kelch domain containing 2 |
| chr17_+_8924837 | 0.07 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr16_+_28858004 | 0.07 |
ENST00000322610.8
|
SH2B1
|
SH2B adaptor protein 1 |
| chr20_+_44035847 | 0.07 |
ENST00000372712.2
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr2_-_128615517 | 0.07 |
ENST00000409698.1
|
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr12_-_92539614 | 0.07 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr16_-_28857677 | 0.07 |
ENST00000313511.3
|
TUFM
|
Tu translation elongation factor, mitochondrial |
| chr12_-_71031185 | 0.07 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr11_+_123301012 | 0.07 |
ENST00000533341.1
|
AP000783.1
|
Uncharacterized protein |
| chr14_+_23067146 | 0.07 |
ENST00000428304.2
|
ABHD4
|
abhydrolase domain containing 4 |
| chr5_+_176853702 | 0.07 |
ENST00000507633.1
ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6
|
G protein-coupled receptor kinase 6 |
| chr16_+_3333443 | 0.07 |
ENST00000572748.1
ENST00000573578.1 ENST00000574253.1 |
ZNF263
|
zinc finger protein 263 |
| chr19_+_18111927 | 0.07 |
ENST00000379656.3
|
ARRDC2
|
arrestin domain containing 2 |
| chr20_+_44035200 | 0.07 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr6_+_32121908 | 0.06 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr3_-_49066811 | 0.06 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr22_-_39928823 | 0.06 |
ENST00000334678.3
|
RPS19BP1
|
ribosomal protein S19 binding protein 1 |
| chr17_-_17875688 | 0.06 |
ENST00000379504.3
ENST00000318094.10 ENST00000540946.1 ENST00000542206.1 ENST00000395739.4 ENST00000581396.1 ENST00000535933.1 ENST00000579586.1 |
TOM1L2
|
target of myb1-like 2 (chicken) |
| chr6_-_42016385 | 0.06 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr19_+_17378159 | 0.06 |
ENST00000598188.1
ENST00000359435.4 ENST00000599474.1 ENST00000599057.1 ENST00000601043.1 ENST00000447614.2 |
BABAM1
|
BRISC and BRCA1 A complex member 1 |
| chr19_+_17378278 | 0.06 |
ENST00000596335.1
ENST00000601436.1 ENST00000595632.1 |
BABAM1
|
BRISC and BRCA1 A complex member 1 |
| chr14_+_105267250 | 0.06 |
ENST00000342537.7
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr12_-_111358372 | 0.06 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr8_-_30002179 | 0.06 |
ENST00000320542.3
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4 |
| chr4_-_90756769 | 0.06 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr1_+_43855545 | 0.06 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr6_+_4087664 | 0.06 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr5_-_115872124 | 0.06 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr12_-_100656134 | 0.06 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr11_-_65150103 | 0.06 |
ENST00000294187.6
ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr19_-_40791302 | 0.05 |
ENST00000392038.2
ENST00000578123.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr19_+_13134772 | 0.05 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr1_+_153940346 | 0.05 |
ENST00000405694.3
ENST00000449724.1 ENST00000368607.3 ENST00000271889.4 |
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr11_+_46639071 | 0.05 |
ENST00000580238.1
ENST00000581416.1 ENST00000529655.1 ENST00000533325.1 ENST00000581438.1 ENST00000583249.1 ENST00000530500.1 ENST00000526508.1 ENST00000578626.1 ENST00000577256.1 ENST00000524625.1 ENST00000582547.1 ENST00000359513.4 ENST00000528494.1 |
ATG13
|
autophagy related 13 |
| chr6_+_32121789 | 0.05 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr14_+_32964258 | 0.05 |
ENST00000556638.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr12_+_121416489 | 0.05 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr12_+_96588368 | 0.05 |
ENST00000547860.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr1_-_43855444 | 0.05 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr1_-_43855479 | 0.05 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr12_+_121416340 | 0.05 |
ENST00000257555.6
ENST00000400024.2 |
HNF1A
|
HNF1 homeobox A |
| chr3_-_185826855 | 0.05 |
ENST00000306376.5
|
ETV5
|
ets variant 5 |
| chr10_-_61900762 | 0.05 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr8_+_95558771 | 0.05 |
ENST00000391679.1
|
AC023632.1
|
HCG2009141; PRO2397; Uncharacterized protein |
| chr11_+_77532155 | 0.05 |
ENST00000532481.1
ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr1_-_153931052 | 0.05 |
ENST00000368630.3
ENST00000368633.1 |
CRTC2
|
CREB regulated transcription coactivator 2 |
| chr19_+_13906250 | 0.05 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr12_-_71031220 | 0.05 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr11_+_33061543 | 0.05 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr4_-_159080806 | 0.05 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr6_-_152623231 | 0.05 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr8_-_134309823 | 0.05 |
ENST00000414097.2
|
NDRG1
|
N-myc downstream regulated 1 |
| chr1_+_153940741 | 0.05 |
ENST00000431292.1
|
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr1_-_153514241 | 0.04 |
ENST00000368718.1
ENST00000359215.1 |
S100A5
|
S100 calcium binding protein A5 |
| chr19_-_7293942 | 0.04 |
ENST00000341500.5
ENST00000302850.5 |
INSR
|
insulin receptor |
| chr1_-_31661000 | 0.04 |
ENST00000263693.1
ENST00000398657.2 ENST00000526106.1 |
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr4_-_140223670 | 0.04 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr17_-_67138015 | 0.04 |
ENST00000284425.2
ENST00000590645.1 |
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr6_+_13182751 | 0.04 |
ENST00000415087.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr8_-_134309335 | 0.04 |
ENST00000522890.1
ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1
|
N-myc downstream regulated 1 |
| chr20_-_44993012 | 0.04 |
ENST00000372229.1
ENST00000372230.5 ENST00000543605.1 ENST00000243896.2 ENST00000317734.8 |
SLC35C2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr18_+_77160282 | 0.04 |
ENST00000318065.5
ENST00000545796.1 ENST00000592223.1 ENST00000329101.4 ENST00000586434.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr12_-_49504449 | 0.04 |
ENST00000547675.1
|
LMBR1L
|
limb development membrane protein 1-like |
| chr6_-_152489484 | 0.04 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr1_+_150122034 | 0.04 |
ENST00000025469.6
ENST00000369124.4 |
PLEKHO1
|
pleckstrin homology domain containing, family O member 1 |
| chr7_+_12250943 | 0.04 |
ENST00000442107.1
|
TMEM106B
|
transmembrane protein 106B |
| chr4_-_102268628 | 0.04 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_+_162039558 | 0.04 |
ENST00000530878.1
ENST00000361897.5 |
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr22_-_43398442 | 0.04 |
ENST00000422336.1
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr3_+_101504200 | 0.04 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr1_-_241803649 | 0.04 |
ENST00000366554.2
|
OPN3
|
opsin 3 |
| chr17_+_33474826 | 0.04 |
ENST00000268876.5
ENST00000433649.1 ENST00000378449.1 |
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr19_-_29704448 | 0.04 |
ENST00000304863.4
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr6_-_31830655 | 0.04 |
ENST00000375631.4
|
NEU1
|
sialidase 1 (lysosomal sialidase) |
| chr2_+_66662510 | 0.04 |
ENST00000272369.9
ENST00000407092.2 |
MEIS1
|
Meis homeobox 1 |
| chr20_-_43150601 | 0.04 |
ENST00000541235.1
ENST00000255175.1 ENST00000342374.4 |
SERINC3
|
serine incorporator 3 |
| chr12_+_54378849 | 0.04 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr2_-_219157250 | 0.04 |
ENST00000434015.2
ENST00000444183.1 ENST00000420341.1 ENST00000453281.1 ENST00000258412.3 ENST00000440422.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr20_-_1309809 | 0.04 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chrX_-_110655391 | 0.04 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr14_-_36988882 | 0.04 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr5_+_137203541 | 0.04 |
ENST00000421631.2
|
MYOT
|
myotilin |
| chr3_+_122513642 | 0.04 |
ENST00000261038.5
|
DIRC2
|
disrupted in renal carcinoma 2 |
| chr14_+_24439148 | 0.04 |
ENST00000543805.1
ENST00000534993.1 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr19_+_2270283 | 0.04 |
ENST00000588673.2
|
OAZ1
|
ornithine decarboxylase antizyme 1 |
| chr10_-_4285923 | 0.04 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr22_+_44427230 | 0.03 |
ENST00000444029.1
|
PARVB
|
parvin, beta |
| chr1_-_57045228 | 0.03 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr4_+_15376165 | 0.03 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr19_-_36523529 | 0.03 |
ENST00000593074.1
|
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr5_+_137203557 | 0.03 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chrX_-_110655306 | 0.03 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr10_-_118032979 | 0.03 |
ENST00000355422.6
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr9_-_39288092 | 0.03 |
ENST00000323947.7
ENST00000297668.6 ENST00000377656.2 ENST00000377659.1 |
CNTNAP3
|
contactin associated protein-like 3 |
| chr15_-_52971544 | 0.03 |
ENST00000566768.1
ENST00000561543.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr4_-_100242549 | 0.03 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr3_-_185826718 | 0.03 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr10_-_73848086 | 0.03 |
ENST00000536168.1
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr12_-_56352368 | 0.03 |
ENST00000549404.1
|
PMEL
|
premelanosome protein |
| chr1_+_40840320 | 0.03 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr18_+_60382672 | 0.03 |
ENST00000400316.4
ENST00000262719.5 |
PHLPP1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr4_-_85771168 | 0.03 |
ENST00000514071.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr10_+_99400443 | 0.03 |
ENST00000370631.3
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr10_-_52645416 | 0.03 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr3_+_160394940 | 0.03 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chr1_-_16763685 | 0.03 |
ENST00000540400.1
|
SPATA21
|
spermatogenesis associated 21 |
| chr4_+_74347400 | 0.03 |
ENST00000226355.3
|
AFM
|
afamin |
| chr2_-_71454185 | 0.03 |
ENST00000244221.8
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chrX_-_38080077 | 0.03 |
ENST00000378533.3
ENST00000544439.1 ENST00000432886.2 ENST00000538295.1 |
SRPX
|
sushi-repeat containing protein, X-linked |
| chr2_+_42104692 | 0.03 |
ENST00000398796.2
ENST00000442214.1 |
AC104654.1
|
AC104654.1 |
| chr8_+_31497271 | 0.03 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr1_-_20126365 | 0.03 |
ENST00000294543.6
ENST00000375122.2 |
TMCO4
|
transmembrane and coiled-coil domains 4 |
| chr16_+_84801852 | 0.03 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr1_-_6420737 | 0.03 |
ENST00000541130.1
ENST00000377845.3 |
ACOT7
|
acyl-CoA thioesterase 7 |
| chr8_-_6420759 | 0.03 |
ENST00000523120.1
|
ANGPT2
|
angiopoietin 2 |
| chrX_+_9431324 | 0.03 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr8_+_67405794 | 0.03 |
ENST00000522977.1
ENST00000480005.1 |
C8orf46
|
chromosome 8 open reading frame 46 |
| chr1_+_47533160 | 0.03 |
ENST00000334194.3
|
CYP4Z1
|
cytochrome P450, family 4, subfamily Z, polypeptide 1 |
| chr5_+_162932554 | 0.03 |
ENST00000321757.6
ENST00000421814.2 ENST00000518095.1 |
MAT2B
|
methionine adenosyltransferase II, beta |
| chr11_+_12115543 | 0.03 |
ENST00000537344.1
ENST00000532179.1 ENST00000526065.1 |
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr1_+_73771844 | 0.03 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
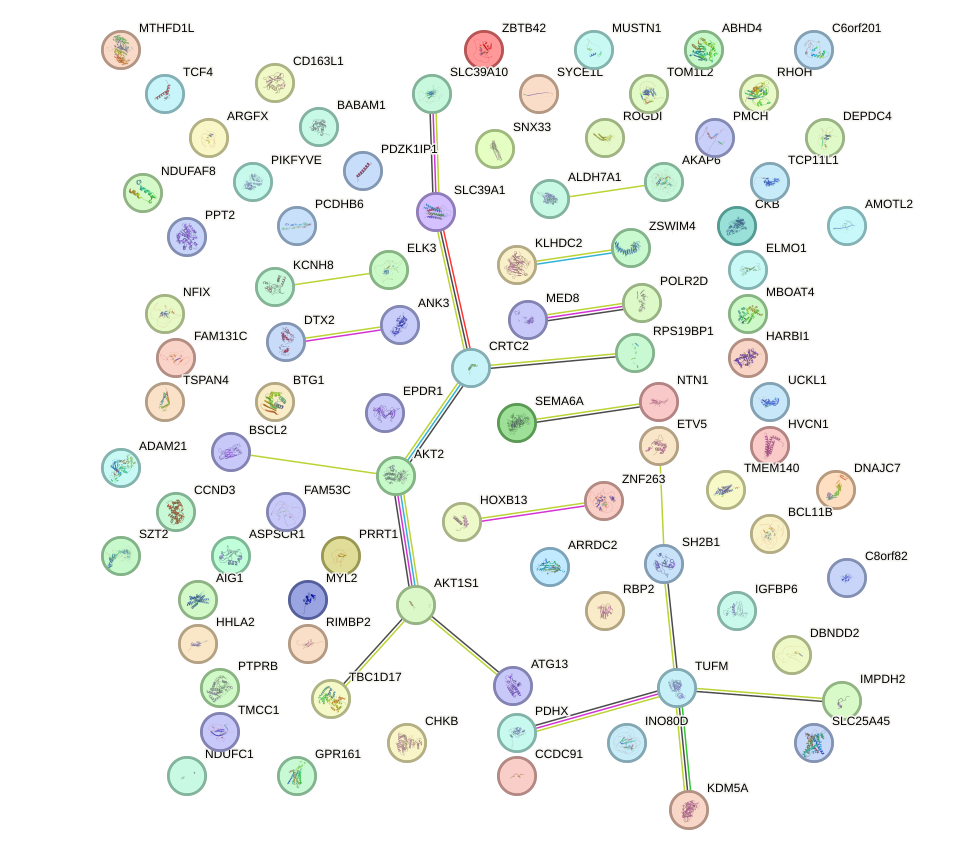
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXO4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.2 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.1 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:1903285 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.2 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.0 | GO:1903762 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.5 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.0 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.0 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.0 | GO:0002276 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.0 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) positive regulation of connective tissue replacement(GO:1905205) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.0 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.0 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.0 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |