Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
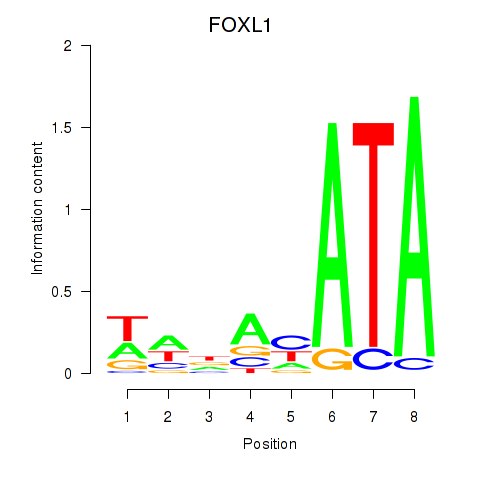
Results for FOXL1
Z-value: 0.75
Transcription factors associated with FOXL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXL1
|
ENSG00000176678.4 | forkhead box L1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXL1 | hg19_v2_chr16_+_86612112_86612123 | -0.73 | 2.7e-01 | Click! |
Activity profile of FOXL1 motif
Sorted Z-values of FOXL1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_238108575 | 0.31 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr1_+_174670143 | 0.29 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr6_+_37012607 | 0.28 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr21_+_35107346 | 0.26 |
ENST00000456489.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr15_-_80263506 | 0.26 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr8_+_52730143 | 0.25 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr18_+_68002675 | 0.24 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr5_-_145483932 | 0.23 |
ENST00000311450.4
|
PLAC8L1
|
PLAC8-like 1 |
| chr9_-_125240235 | 0.23 |
ENST00000259357.2
|
OR1J1
|
olfactory receptor, family 1, subfamily J, member 1 |
| chr19_+_9203855 | 0.23 |
ENST00000429566.3
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1 |
| chr9_-_21077939 | 0.21 |
ENST00000380232.2
|
IFNB1
|
interferon, beta 1, fibroblast |
| chr3_-_25915189 | 0.21 |
ENST00000451284.1
|
LINC00692
|
long intergenic non-protein coding RNA 692 |
| chr17_+_18647326 | 0.21 |
ENST00000395667.1
ENST00000395665.4 ENST00000308799.4 ENST00000301938.4 |
FBXW10
|
F-box and WD repeat domain containing 10 |
| chr12_-_79849240 | 0.20 |
ENST00000550268.1
|
RP1-78O14.1
|
RP1-78O14.1 |
| chr12_+_104337515 | 0.20 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr12_+_20963647 | 0.19 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr12_-_11002063 | 0.18 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr10_-_46342675 | 0.18 |
ENST00000492347.1
|
AGAP4
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 4 |
| chr1_+_95975672 | 0.18 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr2_-_74007193 | 0.18 |
ENST00000377706.4
ENST00000443070.1 ENST00000272444.3 |
DUSP11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr7_+_116654935 | 0.18 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr9_-_95244781 | 0.18 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr21_+_25801041 | 0.18 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr14_+_104177607 | 0.17 |
ENST00000429169.1
|
AL049840.1
|
Uncharacterized protein; cDNA FLJ53535 |
| chr6_-_134861089 | 0.17 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr3_+_57882061 | 0.17 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chr3_+_193853927 | 0.17 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr19_+_20959098 | 0.17 |
ENST00000360204.5
ENST00000594534.1 |
ZNF66
|
zinc finger protein 66 |
| chr4_-_84035905 | 0.17 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr10_-_90967063 | 0.16 |
ENST00000371852.2
|
CH25H
|
cholesterol 25-hydroxylase |
| chr12_+_42624050 | 0.16 |
ENST00000601185.1
|
AC020629.1
|
Uncharacterized protein |
| chr6_+_76599809 | 0.16 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr3_-_93747425 | 0.16 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr1_+_145525015 | 0.16 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr17_+_22022437 | 0.16 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr2_-_151395525 | 0.16 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr8_+_67405438 | 0.15 |
ENST00000305454.3
|
C8orf46
|
chromosome 8 open reading frame 46 |
| chr12_-_100486668 | 0.15 |
ENST00000550544.1
ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L
|
UHRF1 binding protein 1-like |
| chr6_+_153552455 | 0.15 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr8_+_71485681 | 0.15 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1 |
| chr4_+_165675197 | 0.15 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr11_+_112047087 | 0.15 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr3_-_185641681 | 0.15 |
ENST00000259043.7
|
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr13_+_53030107 | 0.15 |
ENST00000490903.1
ENST00000480747.1 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr18_+_158327 | 0.15 |
ENST00000582707.1
|
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) |
| chr4_-_74904398 | 0.15 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr2_-_151344172 | 0.14 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr4_+_76439095 | 0.14 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr6_+_3982909 | 0.14 |
ENST00000356722.3
|
C6ORF50
|
C6ORF50 |
| chr10_-_69597828 | 0.14 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chrX_-_139866723 | 0.14 |
ENST00000370532.2
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa |
| chr12_-_8088773 | 0.14 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr13_+_111855414 | 0.14 |
ENST00000375737.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr15_+_63414760 | 0.14 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr2_-_99797390 | 0.14 |
ENST00000422537.2
|
MITD1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr5_-_95550754 | 0.14 |
ENST00000502437.1
|
RP11-254I22.3
|
RP11-254I22.3 |
| chr9_-_140142222 | 0.14 |
ENST00000344774.4
ENST00000388932.2 |
FAM166A
|
family with sequence similarity 166, member A |
| chr7_+_16793160 | 0.14 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr4_-_39367949 | 0.14 |
ENST00000503784.1
ENST00000349703.2 ENST00000381897.1 |
RFC1
|
replication factor C (activator 1) 1, 145kDa |
| chr1_-_247171347 | 0.14 |
ENST00000339986.7
ENST00000487338.2 |
ZNF695
|
zinc finger protein 695 |
| chr4_-_6694189 | 0.14 |
ENST00000596858.1
|
AC093323.1
|
Uncharacterized protein |
| chr4_+_76871883 | 0.13 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr9_+_135937365 | 0.13 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chr14_+_57671888 | 0.13 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr13_+_37581115 | 0.13 |
ENST00000481013.1
|
EXOSC8
|
exosome component 8 |
| chr6_+_79577189 | 0.13 |
ENST00000369940.2
|
IRAK1BP1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr16_-_69418553 | 0.13 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr10_-_14574705 | 0.13 |
ENST00000489100.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr8_+_109455845 | 0.13 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr1_+_156308403 | 0.13 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr4_-_80994210 | 0.13 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr8_-_117043 | 0.13 |
ENST00000320901.3
|
OR4F21
|
olfactory receptor, family 4, subfamily F, member 21 |
| chr20_+_57594309 | 0.13 |
ENST00000217133.1
|
TUBB1
|
tubulin, beta 1 class VI |
| chr19_+_11877838 | 0.13 |
ENST00000357901.4
ENST00000454339.2 |
ZNF441
|
zinc finger protein 441 |
| chr1_-_227505289 | 0.13 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr5_+_81601166 | 0.13 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr20_-_14318248 | 0.13 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr20_-_33264886 | 0.13 |
ENST00000217446.3
ENST00000452740.2 ENST00000374820.2 |
PIGU
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr2_-_4703793 | 0.12 |
ENST00000421212.1
ENST00000412134.1 |
AC022311.1
|
AC022311.1 |
| chr5_-_78808617 | 0.12 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr15_+_23255242 | 0.12 |
ENST00000450802.3
|
GOLGA8I
|
golgin A8 family, member I |
| chr10_-_14614311 | 0.12 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr14_+_104029362 | 0.12 |
ENST00000495778.1
|
APOPT1
|
apoptogenic 1, mitochondrial |
| chr5_+_179135246 | 0.12 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr6_+_24775153 | 0.12 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr12_-_14849470 | 0.12 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr20_-_35274548 | 0.12 |
ENST00000262866.4
|
SLA2
|
Src-like-adaptor 2 |
| chr12_-_21757774 | 0.12 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver) |
| chr8_-_82598067 | 0.12 |
ENST00000523942.1
ENST00000522997.1 |
IMPA1
|
inositol(myo)-1(or 4)-monophosphatase 1 |
| chr2_+_108905095 | 0.12 |
ENST00000251481.6
ENST00000326853.5 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr5_-_55777586 | 0.12 |
ENST00000506836.1
|
CTC-236F12.4
|
Uncharacterized protein |
| chr5_+_176692466 | 0.12 |
ENST00000508029.1
ENST00000503056.1 |
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr22_-_32766972 | 0.12 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr17_+_71228740 | 0.12 |
ENST00000268942.8
ENST00000359042.2 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr14_+_56127960 | 0.12 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr10_-_14596140 | 0.12 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr5_+_101569696 | 0.12 |
ENST00000597120.1
|
AC008948.1
|
AC008948.1 |
| chr3_-_148939598 | 0.12 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr12_-_121477039 | 0.12 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr11_+_93479588 | 0.11 |
ENST00000526335.1
|
C11orf54
|
chromosome 11 open reading frame 54 |
| chr1_-_20755140 | 0.11 |
ENST00000418743.1
ENST00000426428.1 |
RP4-749H3.1
|
long intergenic non-protein coding RNA 1141 |
| chr1_-_145382434 | 0.11 |
ENST00000610154.1
|
RP11-458D21.1
|
RP11-458D21.1 |
| chr17_-_67057047 | 0.11 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr7_+_110731062 | 0.11 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr15_+_35270552 | 0.11 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr2_-_45795145 | 0.11 |
ENST00000535761.1
|
SRBD1
|
S1 RNA binding domain 1 |
| chr4_-_84035868 | 0.11 |
ENST00000426923.2
ENST00000509973.1 |
PLAC8
|
placenta-specific 8 |
| chr2_-_170681324 | 0.11 |
ENST00000409340.1
|
METTL5
|
methyltransferase like 5 |
| chrX_-_24045303 | 0.11 |
ENST00000328046.8
|
KLHL15
|
kelch-like family member 15 |
| chr8_+_11666649 | 0.11 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr12_-_110883346 | 0.11 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr1_+_104615595 | 0.11 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr4_-_14889791 | 0.11 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
| chr14_+_77582905 | 0.11 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr16_-_28192360 | 0.11 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chrX_-_55020511 | 0.11 |
ENST00000375006.3
ENST00000374992.2 |
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr14_+_35747825 | 0.11 |
ENST00000540871.1
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr8_-_62559366 | 0.11 |
ENST00000522919.1
|
ASPH
|
aspartate beta-hydroxylase |
| chr7_-_14026063 | 0.11 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr2_+_38177575 | 0.11 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr10_+_35456444 | 0.11 |
ENST00000361599.4
|
CREM
|
cAMP responsive element modulator |
| chr12_+_20963632 | 0.11 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr4_-_89619386 | 0.11 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr2_+_113321939 | 0.11 |
ENST00000458012.2
|
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr5_+_159848854 | 0.11 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr16_-_28374829 | 0.11 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chr12_+_43086018 | 0.11 |
ENST00000550177.1
|
RP11-25I15.3
|
RP11-25I15.3 |
| chr4_+_113066552 | 0.10 |
ENST00000309733.5
|
C4orf32
|
chromosome 4 open reading frame 32 |
| chr1_+_225140372 | 0.10 |
ENST00000366848.1
ENST00000439375.2 |
DNAH14
|
dynein, axonemal, heavy chain 14 |
| chr7_-_55930443 | 0.10 |
ENST00000388975.3
|
SEPT14
|
septin 14 |
| chrX_+_108779004 | 0.10 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr2_-_68547061 | 0.10 |
ENST00000263655.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr3_-_107099454 | 0.10 |
ENST00000593837.1
ENST00000599431.1 |
RP11-446H18.5
|
RP11-446H18.5 |
| chr1_-_207226313 | 0.10 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr4_-_69536346 | 0.10 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr5_-_87980587 | 0.10 |
ENST00000509783.1
ENST00000509405.1 ENST00000506978.1 ENST00000509265.1 ENST00000513805.1 |
LINC00461
|
long intergenic non-protein coding RNA 461 |
| chr3_+_101292939 | 0.10 |
ENST00000265260.3
ENST00000469941.1 ENST00000296024.5 |
PCNP
|
PEST proteolytic signal containing nuclear protein |
| chr18_-_37380230 | 0.10 |
ENST00000591629.1
|
LINC00669
|
long intergenic non-protein coding RNA 669 |
| chr7_-_8302298 | 0.10 |
ENST00000446305.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr10_+_695888 | 0.10 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr1_+_28527070 | 0.10 |
ENST00000596102.1
|
AL353354.2
|
AL353354.2 |
| chr2_+_90458201 | 0.10 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr14_+_52456327 | 0.10 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr2_-_74007095 | 0.10 |
ENST00000452812.1
|
DUSP11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr1_-_86848760 | 0.10 |
ENST00000460698.2
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr4_+_78829479 | 0.10 |
ENST00000504901.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr2_+_201450591 | 0.10 |
ENST00000374700.2
|
AOX1
|
aldehyde oxidase 1 |
| chr1_-_33116128 | 0.10 |
ENST00000436661.1
ENST00000373501.2 ENST00000341885.5 ENST00000468695.1 |
ZBTB8OS
|
zinc finger and BTB domain containing 8 opposite strand |
| chr2_+_65454863 | 0.10 |
ENST00000260641.5
|
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast) |
| chr1_-_36235559 | 0.10 |
ENST00000251195.5
|
CLSPN
|
claspin |
| chr1_+_218683438 | 0.10 |
ENST00000443836.1
|
C1orf143
|
chromosome 1 open reading frame 143 |
| chr14_+_50291993 | 0.10 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr14_-_36645674 | 0.10 |
ENST00000556013.2
|
PTCSC3
|
papillary thyroid carcinoma susceptibility candidate 3 (non-protein coding) |
| chr12_-_122107549 | 0.10 |
ENST00000355329.3
|
MORN3
|
MORN repeat containing 3 |
| chr6_-_52109335 | 0.10 |
ENST00000336123.4
|
IL17F
|
interleukin 17F |
| chr10_+_35484053 | 0.09 |
ENST00000487763.1
ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM
|
cAMP responsive element modulator |
| chr8_-_112248400 | 0.09 |
ENST00000519506.1
ENST00000522778.1 |
RP11-946L20.4
|
RP11-946L20.4 |
| chr7_-_102985035 | 0.09 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr19_+_21203426 | 0.09 |
ENST00000261560.5
ENST00000599548.1 ENST00000594110.1 |
ZNF430
|
zinc finger protein 430 |
| chr15_-_49913126 | 0.09 |
ENST00000561064.1
ENST00000299338.6 |
FAM227B
|
family with sequence similarity 227, member B |
| chr5_-_65018834 | 0.09 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr8_+_93895865 | 0.09 |
ENST00000391681.1
|
AC117834.1
|
AC117834.1 |
| chr6_-_90348440 | 0.09 |
ENST00000520441.1
ENST00000520318.1 ENST00000523377.1 |
LYRM2
|
LYR motif containing 2 |
| chr3_-_98235962 | 0.09 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr12_+_133614062 | 0.09 |
ENST00000540031.1
ENST00000536123.1 |
ZNF84
|
zinc finger protein 84 |
| chr19_+_46531127 | 0.09 |
ENST00000601033.1
|
CTC-344H19.4
|
CTC-344H19.4 |
| chr13_+_23993097 | 0.09 |
ENST00000443092.1
|
SACS-AS1
|
SACS antisense RNA 1 |
| chr14_+_53196872 | 0.09 |
ENST00000442123.2
ENST00000354586.4 |
STYX
|
serine/threonine/tyrosine interacting protein |
| chr1_-_149459549 | 0.09 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr4_+_124571409 | 0.09 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr11_+_69924397 | 0.09 |
ENST00000355303.5
|
ANO1
|
anoctamin 1, calcium activated chloride channel |
| chr8_-_101734170 | 0.09 |
ENST00000522387.1
ENST00000518196.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_+_207943667 | 0.09 |
ENST00000462968.2
|
CD46
|
CD46 molecule, complement regulatory protein |
| chr8_+_66619277 | 0.09 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr2_+_62132781 | 0.09 |
ENST00000311832.5
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr9_-_100707116 | 0.09 |
ENST00000259456.3
|
HEMGN
|
hemogen |
| chr9_-_47314222 | 0.09 |
ENST00000420228.1
ENST00000438517.1 ENST00000414020.1 |
AL953854.2
|
AL953854.2 |
| chr6_-_112408661 | 0.09 |
ENST00000368662.5
|
TUBE1
|
tubulin, epsilon 1 |
| chr8_+_94710789 | 0.09 |
ENST00000523475.1
|
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr4_-_80994619 | 0.09 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr2_+_29001711 | 0.09 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr14_-_92198403 | 0.09 |
ENST00000553329.1
ENST00000256343.3 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr2_+_149447783 | 0.09 |
ENST00000449013.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chrM_+_12331 | 0.09 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr17_-_62499334 | 0.09 |
ENST00000579996.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr12_-_121476959 | 0.09 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr10_+_5488564 | 0.09 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr8_+_26247878 | 0.09 |
ENST00000518611.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr14_-_50319758 | 0.09 |
ENST00000298310.5
|
NEMF
|
nuclear export mediator factor |
| chr9_-_98268883 | 0.09 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr1_-_198906528 | 0.09 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr1_-_36235529 | 0.09 |
ENST00000318121.3
ENST00000373220.3 ENST00000520551.1 |
CLSPN
|
claspin |
| chr1_+_160709055 | 0.09 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr17_+_71228793 | 0.09 |
ENST00000426147.2
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chr5_+_96077888 | 0.09 |
ENST00000509259.1
ENST00000503828.1 |
CAST
|
calpastatin |
| chr1_+_145524891 | 0.08 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr12_-_102455846 | 0.08 |
ENST00000545679.1
|
CCDC53
|
coiled-coil domain containing 53 |
| chr6_-_132272504 | 0.08 |
ENST00000367976.3
|
CTGF
|
connective tissue growth factor |
| chr5_-_157286104 | 0.08 |
ENST00000530742.1
ENST00000523908.1 ENST00000523094.1 ENST00000296951.5 ENST00000411809.2 |
CLINT1
|
clathrin interactor 1 |
| chr11_-_3400330 | 0.08 |
ENST00000427810.2
ENST00000005082.9 ENST00000534569.1 ENST00000438262.2 ENST00000528796.1 ENST00000528410.1 ENST00000529678.1 ENST00000354599.6 ENST00000526601.1 ENST00000525502.1 ENST00000533036.1 ENST00000399602.4 |
ZNF195
|
zinc finger protein 195 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXL1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.1 | 0.2 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.2 | GO:0060164 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) regulation of timing of neuron differentiation(GO:0060164) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.1 | 0.2 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.2 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.0 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.2 | GO:0071042 | nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.2 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:1903770 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0033488 | cholesterol biosynthetic process via 24,25-dihydrolanosterol(GO:0033488) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 1.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.1 | GO:0045423 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.2 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0042704 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) uterine wall breakdown(GO:0042704) positive regulation of catagen(GO:0051795) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.0 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.0 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.0 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.2 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.0 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.0 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.0 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.0 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.0 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.0 | 0.1 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.0 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.0 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.0 | GO:0042253 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.0 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.0 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.0 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.0 | GO:1903423 | synaptic vesicle recycling via endosome(GO:0036466) positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.0 | GO:0002519 | natural killer cell tolerance induction(GO:0002519) regulation of tolerance induction dependent upon immune response(GO:0002652) |
| 0.0 | 0.1 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.0 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.0 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.0 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.1 | 0.2 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0008398 | sterol 14-demethylase activity(GO:0008398) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 1.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.0 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.0 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.0 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.0 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.0 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |