Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
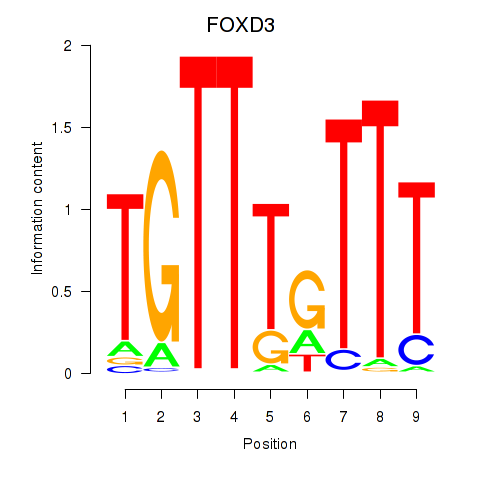
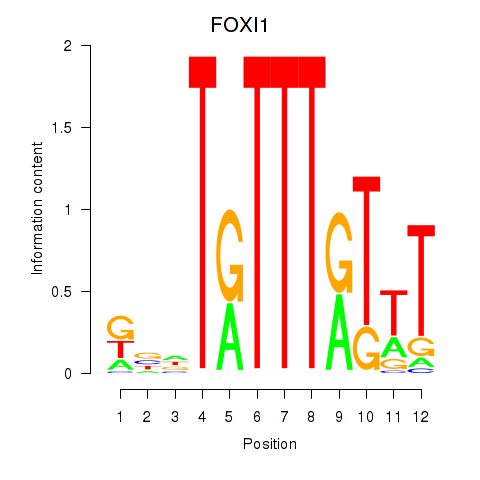
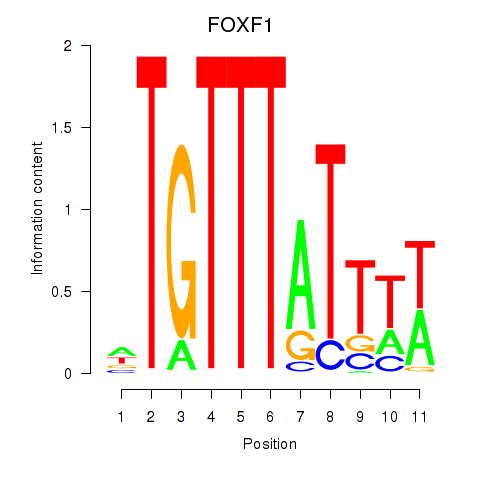
Results for FOXD3_FOXI1_FOXF1
Z-value: 0.98
Transcription factors associated with FOXD3_FOXI1_FOXF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXD3
|
ENSG00000187140.4 | forkhead box D3 |
|
FOXI1
|
ENSG00000168269.7 | forkhead box I1 |
|
FOXF1
|
ENSG00000103241.5 | forkhead box F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXF1 | hg19_v2_chr16_+_86544113_86544145 | 0.82 | 1.8e-01 | Click! |
| FOXD3 | hg19_v2_chr1_+_63788730_63788730 | 0.33 | 6.7e-01 | Click! |
Activity profile of FOXD3_FOXI1_FOXF1 motif
Sorted Z-values of FOXD3_FOXI1_FOXF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_195413160 | 0.68 |
ENST00000599448.1
|
LINC00969
|
long intergenic non-protein coding RNA 969 |
| chr11_-_93271058 | 0.66 |
ENST00000527149.1
|
SMCO4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr5_+_72509751 | 0.64 |
ENST00000515556.1
ENST00000513379.1 ENST00000427584.2 |
RP11-60A8.1
|
RP11-60A8.1 |
| chr7_+_106415457 | 0.49 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr7_-_75401513 | 0.47 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr8_+_97597148 | 0.46 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr11_+_8040739 | 0.43 |
ENST00000534099.1
|
TUB
|
tubby bipartite transcription factor |
| chr2_-_208030295 | 0.42 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr3_-_195310802 | 0.41 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr3_+_136649311 | 0.41 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr6_-_134861089 | 0.40 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr10_-_14050522 | 0.38 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr3_+_57882024 | 0.37 |
ENST00000494088.1
|
SLMAP
|
sarcolemma associated protein |
| chr5_+_40841410 | 0.36 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr10_-_4720301 | 0.34 |
ENST00000449712.1
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr17_-_36358166 | 0.34 |
ENST00000537432.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr4_+_76871883 | 0.34 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr18_-_52626622 | 0.33 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr1_+_95616933 | 0.33 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr9_+_78505581 | 0.32 |
ENST00000376767.3
ENST00000376752.4 |
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr7_+_151038850 | 0.31 |
ENST00000355851.4
ENST00000566856.1 ENST00000470229.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr7_+_134430212 | 0.30 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr6_+_53794780 | 0.30 |
ENST00000505762.1
ENST00000511369.1 ENST00000431554.2 |
MLIP
RP11-411K7.1
|
muscular LMNA-interacting protein RP11-411K7.1 |
| chr4_-_99850243 | 0.30 |
ENST00000280892.6
ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr17_+_79369249 | 0.29 |
ENST00000574717.2
|
RP11-1055B8.6
|
Uncharacterized protein |
| chr9_-_20382446 | 0.28 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr10_-_31288398 | 0.28 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr10_-_128210005 | 0.28 |
ENST00000284694.7
ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr4_+_155484103 | 0.27 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr6_+_4773205 | 0.27 |
ENST00000440139.1
|
CDYL
|
chromodomain protein, Y-like |
| chr12_+_27849378 | 0.27 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr7_+_151791074 | 0.27 |
ENST00000447796.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr4_-_102268708 | 0.26 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr7_-_27219849 | 0.26 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr3_-_121448791 | 0.26 |
ENST00000489400.1
|
GOLGB1
|
golgin B1 |
| chr6_-_131291572 | 0.26 |
ENST00000529208.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr10_-_4720333 | 0.26 |
ENST00000430998.2
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr13_+_52598827 | 0.26 |
ENST00000521776.2
|
UTP14C
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast) |
| chr12_+_25348186 | 0.26 |
ENST00000555711.1
ENST00000556885.1 ENST00000554266.1 ENST00000556351.1 ENST00000556927.1 ENST00000556402.1 ENST00000553788.1 |
LYRM5
|
LYR motif containing 5 |
| chr6_-_131211534 | 0.25 |
ENST00000456097.2
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr7_+_151791037 | 0.25 |
ENST00000419245.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr15_-_55790515 | 0.25 |
ENST00000448430.2
ENST00000457155.2 |
DYX1C1
|
dyslexia susceptibility 1 candidate 1 |
| chr1_+_246729815 | 0.24 |
ENST00000366511.1
|
CNST
|
consortin, connexin sorting protein |
| chr14_+_51026926 | 0.24 |
ENST00000557735.1
|
ATL1
|
atlastin GTPase 1 |
| chr3_-_71179988 | 0.24 |
ENST00000491238.1
|
FOXP1
|
forkhead box P1 |
| chr12_+_25348139 | 0.24 |
ENST00000557540.2
ENST00000381356.4 |
LYRM5
|
LYR motif containing 5 |
| chr4_-_6694189 | 0.24 |
ENST00000596858.1
|
AC093323.1
|
Uncharacterized protein |
| chr2_+_232575128 | 0.24 |
ENST00000412128.1
|
PTMA
|
prothymosin, alpha |
| chr17_-_19290483 | 0.23 |
ENST00000395592.2
ENST00000299610.4 |
MFAP4
|
microfibrillar-associated protein 4 |
| chr1_+_239882842 | 0.23 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr3_+_171561127 | 0.23 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr10_+_114709999 | 0.22 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr4_+_75174204 | 0.22 |
ENST00000332112.4
ENST00000514968.1 ENST00000503098.1 ENST00000502358.1 ENST00000509145.1 ENST00000505212.1 |
EPGN
|
epithelial mitogen |
| chr4_+_71588372 | 0.22 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr4_+_71587669 | 0.22 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr3_-_57233966 | 0.22 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr17_-_37764128 | 0.22 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr4_+_184365744 | 0.21 |
ENST00000504169.1
ENST00000302350.4 |
CDKN2AIP
|
CDKN2A interacting protein |
| chr15_+_63188009 | 0.21 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr4_+_147096837 | 0.21 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr7_+_120591170 | 0.20 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family, member 3 |
| chr3_-_193096600 | 0.20 |
ENST00000446087.1
ENST00000342358.4 |
ATP13A5
|
ATPase type 13A5 |
| chr6_-_107235331 | 0.20 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr19_+_44529479 | 0.20 |
ENST00000587846.1
ENST00000187879.8 ENST00000391960.3 |
ZNF222
|
zinc finger protein 222 |
| chr12_-_68696652 | 0.20 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr12_-_71551652 | 0.20 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr5_-_133510456 | 0.19 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr3_+_157827841 | 0.19 |
ENST00000295930.3
ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr9_+_26956474 | 0.19 |
ENST00000429045.2
|
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr10_-_46030787 | 0.19 |
ENST00000395769.2
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr8_+_107593198 | 0.19 |
ENST00000517686.1
|
OXR1
|
oxidation resistance 1 |
| chr14_+_31046959 | 0.19 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr8_-_123793048 | 0.19 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr6_+_47666275 | 0.19 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr7_-_140624499 | 0.19 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr4_+_155484155 | 0.19 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr16_-_53737722 | 0.19 |
ENST00000569716.1
ENST00000562588.1 ENST00000562230.1 ENST00000379925.3 ENST00000563746.1 ENST00000568653.3 |
RPGRIP1L
|
RPGRIP1-like |
| chr2_+_234294585 | 0.18 |
ENST00000447484.1
|
DGKD
|
diacylglycerol kinase, delta 130kDa |
| chr3_-_123123407 | 0.18 |
ENST00000466617.1
|
ADCY5
|
adenylate cyclase 5 |
| chr2_+_109223595 | 0.18 |
ENST00000410093.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr17_+_39346139 | 0.18 |
ENST00000398470.1
ENST00000318329.5 |
KRTAP9-1
|
keratin associated protein 9-1 |
| chr2_-_111291587 | 0.18 |
ENST00000437167.1
|
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr14_+_56127960 | 0.18 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr11_-_46142615 | 0.18 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr17_+_7533439 | 0.18 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr2_-_152146385 | 0.18 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr17_-_46692457 | 0.17 |
ENST00000468443.1
|
HOXB8
|
homeobox B8 |
| chr6_+_21666633 | 0.17 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr11_-_11374904 | 0.17 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr11_+_34654011 | 0.17 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr17_-_49021974 | 0.17 |
ENST00000501718.2
|
RP11-700H6.1
|
RP11-700H6.1 |
| chr1_-_161207986 | 0.17 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_-_98510843 | 0.17 |
ENST00000413670.2
ENST00000538428.1 |
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr7_+_128312346 | 0.17 |
ENST00000480462.1
ENST00000378704.3 ENST00000477515.1 |
FAM71F2
|
family with sequence similarity 71, member F2 |
| chr4_-_147442817 | 0.16 |
ENST00000507030.1
|
SLC10A7
|
solute carrier family 10, member 7 |
| chr2_+_120517717 | 0.16 |
ENST00000420482.1
ENST00000488279.2 |
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr22_+_21128167 | 0.16 |
ENST00000215727.5
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr9_-_86432547 | 0.16 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr4_+_130014836 | 0.16 |
ENST00000502887.1
|
C4orf33
|
chromosome 4 open reading frame 33 |
| chr14_+_31028348 | 0.16 |
ENST00000550944.1
ENST00000438909.2 ENST00000553504.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr12_-_121476959 | 0.16 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr14_+_39944025 | 0.16 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr11_+_17316870 | 0.16 |
ENST00000458064.2
|
NUCB2
|
nucleobindin 2 |
| chr8_-_42360015 | 0.16 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr1_-_150669604 | 0.16 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr10_+_135340859 | 0.16 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr3_-_71294304 | 0.16 |
ENST00000498215.1
|
FOXP1
|
forkhead box P1 |
| chr16_+_22517166 | 0.15 |
ENST00000356156.3
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr18_+_56806701 | 0.15 |
ENST00000587834.1
|
SEC11C
|
SEC11 homolog C (S. cerevisiae) |
| chrX_-_38186775 | 0.15 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr11_+_73498973 | 0.15 |
ENST00000537007.1
|
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr2_-_69180083 | 0.15 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr22_+_46449674 | 0.15 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr3_+_119316721 | 0.15 |
ENST00000488919.1
ENST00000495992.1 |
PLA1A
|
phospholipase A1 member A |
| chr1_+_174844645 | 0.15 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr3_-_145940126 | 0.15 |
ENST00000498625.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr7_-_83824449 | 0.15 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_+_70026795 | 0.15 |
ENST00000472655.2
ENST00000538810.1 |
RP11-84E24.3
|
long intergenic non-protein coding RNA 1152 |
| chrX_-_38186811 | 0.15 |
ENST00000318842.7
|
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr10_-_94301107 | 0.15 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr4_+_169013666 | 0.14 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chrX_-_70331298 | 0.14 |
ENST00000456850.2
ENST00000473378.1 ENST00000487883.1 ENST00000374202.2 |
IL2RG
|
interleukin 2 receptor, gamma |
| chr11_-_93276582 | 0.14 |
ENST00000298966.2
|
SMCO4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr1_-_227505289 | 0.14 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr6_-_94129244 | 0.14 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chr5_-_38557561 | 0.14 |
ENST00000511561.1
|
LIFR
|
leukemia inhibitory factor receptor alpha |
| chr3_-_71632894 | 0.14 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr11_+_108093755 | 0.14 |
ENST00000527891.1
ENST00000532931.1 |
ATM
|
ataxia telangiectasia mutated |
| chr14_+_77843459 | 0.14 |
ENST00000216471.4
|
SAMD15
|
sterile alpha motif domain containing 15 |
| chr2_-_202222091 | 0.14 |
ENST00000405148.2
ENST00000392257.3 ENST00000439709.1 |
ALS2CR12
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 12 |
| chr3_-_186080012 | 0.14 |
ENST00000544847.1
ENST00000265022.3 |
DGKG
|
diacylglycerol kinase, gamma 90kDa |
| chr5_-_41510725 | 0.14 |
ENST00000328457.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr4_+_124317940 | 0.14 |
ENST00000505319.1
ENST00000339241.1 |
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr4_+_79475019 | 0.14 |
ENST00000508214.1
|
ANXA3
|
annexin A3 |
| chr5_-_65018834 | 0.14 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr13_+_95364963 | 0.13 |
ENST00000438290.2
|
SOX21-AS1
|
SOX21 antisense RNA 1 (head to head) |
| chr5_-_142065612 | 0.13 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr7_+_7811992 | 0.13 |
ENST00000406829.1
|
RPA3-AS1
|
RPA3 antisense RNA 1 |
| chr8_+_104384616 | 0.13 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr11_+_101918153 | 0.13 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr19_-_52531600 | 0.13 |
ENST00000356322.6
ENST00000270649.6 |
ZNF614
|
zinc finger protein 614 |
| chr14_+_61654271 | 0.13 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr7_-_14029283 | 0.13 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr2_+_135596180 | 0.13 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr12_-_113658826 | 0.13 |
ENST00000546692.1
|
IQCD
|
IQ motif containing D |
| chr4_+_103790462 | 0.13 |
ENST00000503643.1
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chr15_-_30114622 | 0.13 |
ENST00000495972.2
ENST00000346128.6 |
TJP1
|
tight junction protein 1 |
| chr2_-_113012592 | 0.13 |
ENST00000272570.5
ENST00000409573.2 |
ZC3H8
|
zinc finger CCCH-type containing 8 |
| chr12_+_29302023 | 0.13 |
ENST00000551451.1
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr9_-_37384431 | 0.13 |
ENST00000452923.1
|
RP11-397D12.4
|
RP11-397D12.4 |
| chr14_+_20215587 | 0.13 |
ENST00000331723.1
|
OR4Q3
|
olfactory receptor, family 4, subfamily Q, member 3 |
| chr12_-_10826612 | 0.13 |
ENST00000535345.1
ENST00000542562.1 |
STYK1
|
serine/threonine/tyrosine kinase 1 |
| chr1_-_243349684 | 0.13 |
ENST00000522895.1
|
CEP170
|
centrosomal protein 170kDa |
| chr20_+_9049303 | 0.13 |
ENST00000407043.2
ENST00000441846.1 |
PLCB4
|
phospholipase C, beta 4 |
| chr12_-_122018114 | 0.13 |
ENST00000539394.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr14_-_92572894 | 0.12 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
| chr12_+_79439405 | 0.12 |
ENST00000552744.1
|
SYT1
|
synaptotagmin I |
| chr1_-_165667545 | 0.12 |
ENST00000538148.1
|
ALDH9A1
|
aldehyde dehydrogenase 9 family, member A1 |
| chr12_-_68845417 | 0.12 |
ENST00000542875.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr4_+_26344754 | 0.12 |
ENST00000515573.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr20_-_5093399 | 0.12 |
ENST00000379276.4
|
TMEM230
|
transmembrane protein 230 |
| chr7_+_151038785 | 0.12 |
ENST00000413040.2
ENST00000568733.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr10_+_111967345 | 0.12 |
ENST00000332674.5
ENST00000453116.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr15_-_38519066 | 0.12 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chr13_+_28712614 | 0.12 |
ENST00000380958.3
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr3_-_57530051 | 0.12 |
ENST00000311202.6
ENST00000351747.2 ENST00000495027.1 ENST00000389536.4 |
DNAH12
|
dynein, axonemal, heavy chain 12 |
| chr12_+_123942188 | 0.12 |
ENST00000526639.2
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr4_+_57774042 | 0.12 |
ENST00000309042.7
|
REST
|
RE1-silencing transcription factor |
| chr10_-_63995871 | 0.12 |
ENST00000315289.2
|
RTKN2
|
rhotekin 2 |
| chr1_-_31666767 | 0.12 |
ENST00000530145.1
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr17_+_73642315 | 0.12 |
ENST00000556126.2
|
SMIM6
|
small integral membrane protein 6 |
| chr17_+_9745786 | 0.12 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr12_-_13529594 | 0.12 |
ENST00000539026.1
|
C12orf36
|
chromosome 12 open reading frame 36 |
| chr17_-_9725388 | 0.12 |
ENST00000399363.4
|
RP11-477N12.3
|
Putative germ cell-specific gene 1-like protein 2 |
| chr15_+_63335899 | 0.12 |
ENST00000561266.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr8_-_28347737 | 0.11 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr1_+_164529004 | 0.11 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr10_+_114710211 | 0.11 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr15_+_76352178 | 0.11 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr2_+_70121075 | 0.11 |
ENST00000409116.1
|
SNRNP27
|
small nuclear ribonucleoprotein 27kDa (U4/U6.U5) |
| chrX_-_154493791 | 0.11 |
ENST00000369454.3
|
RAB39B
|
RAB39B, member RAS oncogene family |
| chr5_+_179135246 | 0.11 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr11_-_8892464 | 0.11 |
ENST00000527347.1
ENST00000526241.1 ENST00000526126.1 ENST00000530938.1 ENST00000526057.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr2_+_136343943 | 0.11 |
ENST00000409606.1
|
R3HDM1
|
R3H domain containing 1 |
| chr15_+_80351977 | 0.11 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr5_-_41510656 | 0.11 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr8_-_117886732 | 0.11 |
ENST00000517485.1
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr3_+_164924716 | 0.11 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr14_+_51026743 | 0.11 |
ENST00000358385.6
ENST00000357032.3 ENST00000354525.4 |
ATL1
|
atlastin GTPase 1 |
| chr10_+_111985837 | 0.11 |
ENST00000393134.1
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr20_+_9049682 | 0.11 |
ENST00000334005.3
ENST00000378473.3 |
PLCB4
|
phospholipase C, beta 4 |
| chr5_+_130506629 | 0.11 |
ENST00000510516.1
ENST00000507584.1 |
LYRM7
|
LYR motif containing 7 |
| chr3_-_121467983 | 0.11 |
ENST00000472475.1
|
GOLGB1
|
golgin B1 |
| chrX_+_155227246 | 0.11 |
ENST00000244174.5
ENST00000424344.3 |
IL9R
|
interleukin 9 receptor |
| chr10_-_61122220 | 0.11 |
ENST00000422313.2
ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C
|
family with sequence similarity 13, member C |
| chr9_+_82267508 | 0.11 |
ENST00000490347.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr10_+_112631699 | 0.11 |
ENST00000444997.1
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor) |
| chr9_+_134103496 | 0.11 |
ENST00000498010.1
ENST00000476004.1 ENST00000528406.1 |
NUP214
|
nucleoporin 214kDa |
| chr9_+_26956371 | 0.11 |
ENST00000380062.5
ENST00000518614.1 |
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr4_+_26321284 | 0.11 |
ENST00000506956.1
ENST00000512671.1 ENST00000345843.3 ENST00000342295.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXD3_FOXI1_FOXF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.7 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.4 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.1 | 0.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.5 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.3 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.1 | 0.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.1 | 0.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.4 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.3 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.2 | GO:0001999 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) renin secretion into blood stream(GO:0002001) |
| 0.0 | 0.1 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.5 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.2 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.1 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.2 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:1904346 | positive regulation of growth rate(GO:0040010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.2 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.0 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:1990828 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.0 | 0.1 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.1 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.0 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.3 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.2 | GO:0071352 | cellular response to interleukin-2(GO:0071352) |
| 0.0 | 0.4 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.1 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.3 | GO:0071600 | otic vesicle morphogenesis(GO:0071600) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.5 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.0 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0070121 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.1 | GO:0043375 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.3 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.2 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.0 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.0 | GO:0070637 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.3 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.0 | GO:0060733 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.0 | 0.0 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.0 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.0 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.0 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.0 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.0 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.0 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.0 | 0.0 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.0 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.1 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.2 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.3 | GO:1990907 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.3 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.0 | 0.2 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.0 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.3 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 0.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.3 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.1 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.3 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.1 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.1 | GO:0031768 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.0 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.0 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.0 | GO:0036317 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.0 | GO:0036328 | VEGF-C-activated receptor activity(GO:0036328) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.0 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.0 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |