Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
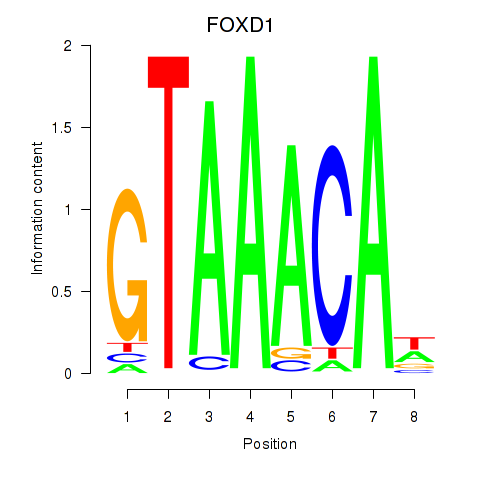
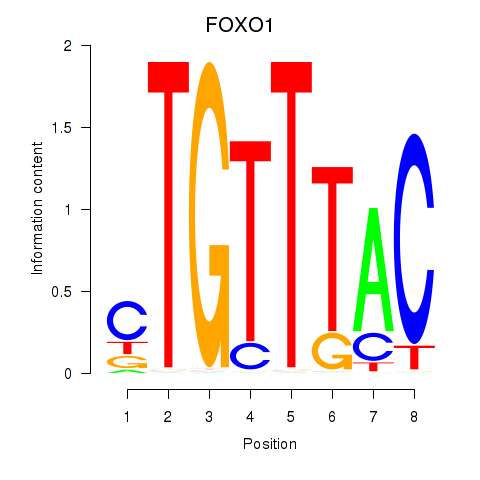
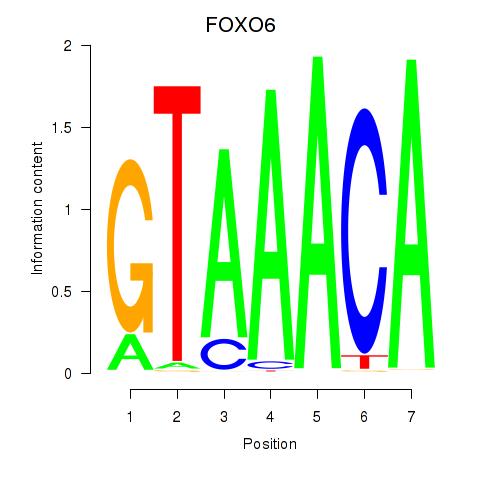
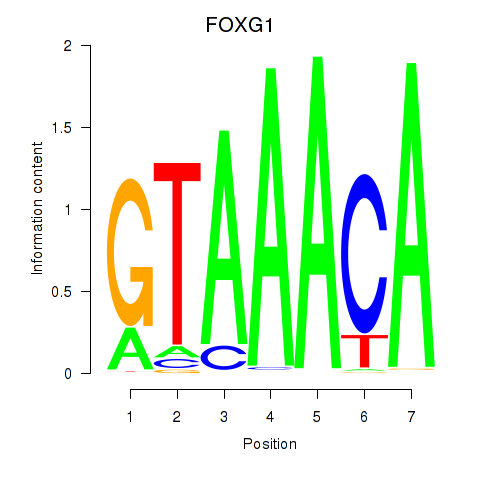
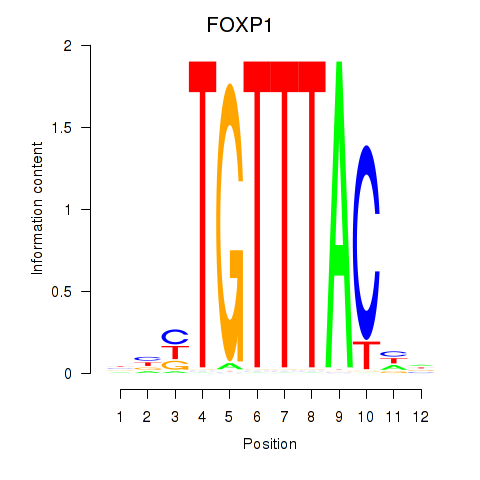
Results for FOXD1_FOXO1_FOXO6_FOXG1_FOXP1
Z-value: 0.25
Transcription factors associated with FOXD1_FOXO1_FOXO6_FOXG1_FOXP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXD1
|
ENSG00000251493.2 | forkhead box D1 |
|
FOXO1
|
ENSG00000150907.6 | forkhead box O1 |
|
FOXO6
|
ENSG00000204060.4 | forkhead box O6 |
|
FOXG1
|
ENSG00000176165.7 | forkhead box G1 |
|
FOXP1
|
ENSG00000114861.14 | forkhead box P1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXO6 | hg19_v2_chr1_+_41827594_41827594 | 0.81 | 1.9e-01 | Click! |
| FOXP1 | hg19_v2_chr3_-_71179699_71179744 | 0.56 | 4.4e-01 | Click! |
| FOXG1 | hg19_v2_chr14_+_29234870_29235050 | 0.52 | 4.8e-01 | Click! |
| FOXD1 | hg19_v2_chr5_-_72744336_72744359 | 0.32 | 6.8e-01 | Click! |
| FOXO1 | hg19_v2_chr13_-_41240717_41240735 | 0.11 | 8.9e-01 | Click! |
Activity profile of FOXD1_FOXO1_FOXO6_FOXG1_FOXP1 motif
Sorted Z-values of FOXD1_FOXO1_FOXO6_FOXG1_FOXP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_79369249 | 0.30 |
ENST00000574717.2
|
RP11-1055B8.6
|
Uncharacterized protein |
| chr16_+_53242350 | 0.29 |
ENST00000565442.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr17_+_9745786 | 0.24 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr1_-_12679171 | 0.23 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr6_-_32122106 | 0.19 |
ENST00000428778.1
|
PRRT1
|
proline-rich transmembrane protein 1 |
| chr15_+_101417919 | 0.19 |
ENST00000561338.1
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr8_-_131028782 | 0.16 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr10_+_43932553 | 0.16 |
ENST00000456416.1
ENST00000437590.2 ENST00000451167.1 |
ZNF487
|
zinc finger protein 487 |
| chr14_-_23288930 | 0.15 |
ENST00000554517.1
ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr7_+_91570240 | 0.14 |
ENST00000394564.1
|
AKAP9
|
A kinase (PRKA) anchor protein 9 |
| chr19_-_47734448 | 0.12 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr3_+_181429704 | 0.12 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr18_+_9475585 | 0.12 |
ENST00000585015.1
|
RALBP1
|
ralA binding protein 1 |
| chr4_-_140223614 | 0.12 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr2_-_160472952 | 0.12 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr18_-_2982869 | 0.11 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr14_+_58894404 | 0.11 |
ENST00000554463.1
ENST00000555833.1 |
KIAA0586
|
KIAA0586 |
| chr17_+_26662679 | 0.11 |
ENST00000578158.1
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr16_-_84150410 | 0.11 |
ENST00000569907.1
|
MBTPS1
|
membrane-bound transcription factor peptidase, site 1 |
| chr8_+_97773202 | 0.11 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr1_-_201096312 | 0.11 |
ENST00000449188.2
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chr12_-_49504449 | 0.11 |
ENST00000547675.1
|
LMBR1L
|
limb development membrane protein 1-like |
| chr4_+_183370146 | 0.11 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr17_-_8059638 | 0.11 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr11_+_844406 | 0.10 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr2_+_149402989 | 0.10 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr17_+_58755184 | 0.10 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr6_-_30043539 | 0.10 |
ENST00000376751.3
ENST00000244360.6 |
RNF39
|
ring finger protein 39 |
| chr7_-_84121858 | 0.09 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr12_-_92539614 | 0.09 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr12_+_54378849 | 0.09 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr3_+_187930429 | 0.09 |
ENST00000420410.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr1_-_27816556 | 0.09 |
ENST00000536657.1
|
WASF2
|
WAS protein family, member 2 |
| chr16_+_4896659 | 0.09 |
ENST00000592120.1
|
UBN1
|
ubinuclein 1 |
| chr5_-_142065612 | 0.09 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr14_-_54418598 | 0.08 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr2_+_111878483 | 0.08 |
ENST00000308659.8
ENST00000357757.2 ENST00000393253.2 ENST00000337565.5 ENST00000393256.3 |
BCL2L11
|
BCL2-like 11 (apoptosis facilitator) |
| chrX_-_54070388 | 0.08 |
ENST00000415025.1
|
PHF8
|
PHD finger protein 8 |
| chr20_+_44035200 | 0.08 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr2_+_177502438 | 0.08 |
ENST00000443670.1
|
AC017048.4
|
long intergenic non-protein coding RNA 1117 |
| chr8_+_97773457 | 0.08 |
ENST00000521142.1
|
CPQ
|
carboxypeptidase Q |
| chr5_-_38557561 | 0.08 |
ENST00000511561.1
|
LIFR
|
leukemia inhibitory factor receptor alpha |
| chr17_+_73455788 | 0.08 |
ENST00000581519.1
|
KIAA0195
|
KIAA0195 |
| chr14_+_32476072 | 0.08 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chr4_-_140223670 | 0.07 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr17_-_46657473 | 0.07 |
ENST00000332503.5
|
HOXB4
|
homeobox B4 |
| chr5_+_175288631 | 0.07 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr11_+_33061336 | 0.07 |
ENST00000602733.1
|
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr6_-_56716686 | 0.07 |
ENST00000520645.1
|
DST
|
dystonin |
| chr16_-_30107491 | 0.07 |
ENST00000566134.1
ENST00000565110.1 ENST00000398841.1 ENST00000398838.4 |
YPEL3
|
yippee-like 3 (Drosophila) |
| chr5_-_126409159 | 0.07 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr4_-_40632757 | 0.07 |
ENST00000511902.1
ENST00000505220.1 |
RBM47
|
RNA binding motif protein 47 |
| chr19_+_13134772 | 0.07 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr20_+_306177 | 0.07 |
ENST00000544632.1
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr3_-_107777208 | 0.07 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr2_+_26785409 | 0.07 |
ENST00000329615.3
ENST00000409392.1 |
C2orf70
|
chromosome 2 open reading frame 70 |
| chr17_+_37821593 | 0.07 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr12_+_121416437 | 0.07 |
ENST00000402929.1
ENST00000535955.1 ENST00000538626.1 ENST00000543427.1 |
HNF1A
|
HNF1 homeobox A |
| chr6_-_33168391 | 0.07 |
ENST00000374685.4
ENST00000413614.2 ENST00000374680.3 |
RXRB
|
retinoid X receptor, beta |
| chr14_-_75083313 | 0.07 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr9_-_20382446 | 0.07 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr14_-_69262789 | 0.07 |
ENST00000557022.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr10_+_99079008 | 0.07 |
ENST00000371021.3
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas |
| chr6_-_131211534 | 0.06 |
ENST00000456097.2
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr2_-_87248975 | 0.06 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr3_+_187930491 | 0.06 |
ENST00000443217.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr1_+_198126209 | 0.06 |
ENST00000367383.1
|
NEK7
|
NIMA-related kinase 7 |
| chr7_+_134551583 | 0.06 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr8_-_94753202 | 0.06 |
ENST00000521947.1
|
RBM12B
|
RNA binding motif protein 12B |
| chr11_+_46402482 | 0.06 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr9_-_98269699 | 0.06 |
ENST00000429896.2
|
PTCH1
|
patched 1 |
| chr2_-_190044480 | 0.06 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr14_-_69262916 | 0.06 |
ENST00000553375.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr9_+_136287444 | 0.06 |
ENST00000355699.2
ENST00000356589.2 ENST00000371911.3 |
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr4_-_174256276 | 0.06 |
ENST00000296503.5
|
HMGB2
|
high mobility group box 2 |
| chr2_+_33661382 | 0.06 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr10_-_99094458 | 0.06 |
ENST00000371019.2
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2 |
| chr19_-_42916499 | 0.06 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr17_-_41322332 | 0.06 |
ENST00000590740.1
|
RP11-242D8.1
|
RP11-242D8.1 |
| chr8_-_94752946 | 0.06 |
ENST00000519109.1
|
RBM12B
|
RNA binding motif protein 12B |
| chr14_-_74551096 | 0.06 |
ENST00000350259.4
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr10_+_111985837 | 0.05 |
ENST00000393134.1
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr6_-_113754604 | 0.05 |
ENST00000421737.1
|
RP1-124C6.1
|
RP1-124C6.1 |
| chr1_-_27816641 | 0.05 |
ENST00000430629.2
|
WASF2
|
WAS protein family, member 2 |
| chr8_+_21777243 | 0.05 |
ENST00000521303.1
|
XPO7
|
exportin 7 |
| chr11_-_57089774 | 0.05 |
ENST00000527207.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr8_-_8318847 | 0.05 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr15_-_83240783 | 0.05 |
ENST00000568994.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chrX_+_70316005 | 0.05 |
ENST00000374259.3
|
FOXO4
|
forkhead box O4 |
| chr17_-_1508379 | 0.05 |
ENST00000412517.3
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr1_+_92545862 | 0.05 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr7_-_105332084 | 0.05 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr5_+_137673200 | 0.05 |
ENST00000434981.2
|
FAM53C
|
family with sequence similarity 53, member C |
| chr9_+_131904295 | 0.05 |
ENST00000434095.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr20_+_306221 | 0.05 |
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr6_+_30853002 | 0.05 |
ENST00000421124.2
ENST00000512725.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr20_+_48909240 | 0.05 |
ENST00000371639.3
|
RP11-290F20.1
|
RP11-290F20.1 |
| chr2_+_175260514 | 0.05 |
ENST00000424069.1
ENST00000427038.1 |
SCRN3
|
secernin 3 |
| chr13_-_67802549 | 0.05 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr1_+_43855545 | 0.05 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr17_+_72426891 | 0.05 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr3_-_135915146 | 0.05 |
ENST00000473093.1
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr12_-_122018114 | 0.05 |
ENST00000539394.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr7_-_150946015 | 0.05 |
ENST00000262188.8
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr17_+_42248063 | 0.05 |
ENST00000293414.1
|
ASB16
|
ankyrin repeat and SOCS box containing 16 |
| chr8_-_94753184 | 0.05 |
ENST00000520560.1
|
RBM12B
|
RNA binding motif protein 12B |
| chr6_-_56707943 | 0.04 |
ENST00000370769.4
ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST
|
dystonin |
| chr20_-_22559211 | 0.04 |
ENST00000564492.1
|
LINC00261
|
long intergenic non-protein coding RNA 261 |
| chr15_-_70994612 | 0.04 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr19_+_13906250 | 0.04 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr1_+_200011711 | 0.04 |
ENST00000544748.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr9_+_2159850 | 0.04 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr9_+_131903916 | 0.04 |
ENST00000419582.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr12_+_121416340 | 0.04 |
ENST00000257555.6
ENST00000400024.2 |
HNF1A
|
HNF1 homeobox A |
| chr9_+_118916082 | 0.04 |
ENST00000328252.3
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr12_-_116714564 | 0.04 |
ENST00000548743.1
|
MED13L
|
mediator complex subunit 13-like |
| chr2_-_202298268 | 0.04 |
ENST00000440597.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr20_+_2082494 | 0.04 |
ENST00000246032.3
|
STK35
|
serine/threonine kinase 35 |
| chr11_+_844067 | 0.04 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr18_+_29171689 | 0.04 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr2_-_27434756 | 0.04 |
ENST00000426119.1
ENST00000412471.1 |
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr7_-_5463175 | 0.04 |
ENST00000399537.4
ENST00000430969.1 |
TNRC18
|
trinucleotide repeat containing 18 |
| chr1_+_90460661 | 0.04 |
ENST00000340281.4
ENST00000361911.5 ENST00000370447.3 ENST00000455342.2 |
ZNF326
|
zinc finger protein 326 |
| chr1_+_246729815 | 0.04 |
ENST00000366511.1
|
CNST
|
consortin, connexin sorting protein |
| chr1_+_164600184 | 0.04 |
ENST00000482110.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr5_-_142783365 | 0.04 |
ENST00000508760.1
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr17_-_42580738 | 0.04 |
ENST00000585614.1
ENST00000591680.1 ENST00000434000.1 ENST00000588554.1 ENST00000592154.1 |
GPATCH8
|
G patch domain containing 8 |
| chr19_-_45909585 | 0.04 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr16_+_4421841 | 0.04 |
ENST00000304735.3
|
VASN
|
vasorin |
| chr17_-_7082668 | 0.04 |
ENST00000573083.1
ENST00000574388.1 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr5_+_40841410 | 0.04 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr5_-_64920115 | 0.04 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr4_+_76871883 | 0.04 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr11_-_115375107 | 0.04 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr12_+_121416489 | 0.04 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr7_+_28448995 | 0.04 |
ENST00000424599.1
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr6_-_42418999 | 0.04 |
ENST00000340840.2
ENST00000354325.2 |
TRERF1
|
transcriptional regulating factor 1 |
| chr2_-_183387064 | 0.04 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr16_-_87729753 | 0.04 |
ENST00000538868.1
|
AC010536.1
|
Uncharacterized protein; cDNA FLJ45526 fis, clone BRTHA2027227 |
| chr1_+_84630367 | 0.04 |
ENST00000370680.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_-_160231451 | 0.04 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr19_-_6057282 | 0.04 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr13_-_23490508 | 0.04 |
ENST00000577004.1
|
LINC00621
|
long intergenic non-protein coding RNA 621 |
| chr17_-_46035187 | 0.04 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr2_-_71454185 | 0.04 |
ENST00000244221.8
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr6_+_144980954 | 0.04 |
ENST00000367525.3
|
UTRN
|
utrophin |
| chr5_+_102200948 | 0.04 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr14_+_21566980 | 0.04 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr5_-_42811986 | 0.04 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr14_-_37051798 | 0.04 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr1_+_40862501 | 0.04 |
ENST00000539317.1
|
SMAP2
|
small ArfGAP2 |
| chr1_+_12806141 | 0.03 |
ENST00000288048.5
|
C1orf158
|
chromosome 1 open reading frame 158 |
| chr3_-_178865747 | 0.03 |
ENST00000435560.1
|
RP11-360P21.2
|
RP11-360P21.2 |
| chr19_+_16435625 | 0.03 |
ENST00000248071.5
ENST00000592003.1 |
KLF2
|
Kruppel-like factor 2 |
| chr7_-_27219849 | 0.03 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr12_-_28125638 | 0.03 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chr16_-_4664860 | 0.03 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr4_-_40632881 | 0.03 |
ENST00000511598.1
|
RBM47
|
RNA binding motif protein 47 |
| chr15_-_34630234 | 0.03 |
ENST00000558667.1
ENST00000561120.1 ENST00000559236.1 ENST00000397702.2 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr2_+_162087577 | 0.03 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chrX_-_106960285 | 0.03 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr14_+_24779376 | 0.03 |
ENST00000530080.1
|
LTB4R2
|
leukotriene B4 receptor 2 |
| chr7_-_100076765 | 0.03 |
ENST00000393991.1
|
TSC22D4
|
TSC22 domain family, member 4 |
| chr4_-_69215699 | 0.03 |
ENST00000510746.1
ENST00000344157.4 ENST00000355665.3 |
YTHDC1
|
YTH domain containing 1 |
| chr4_+_87857538 | 0.03 |
ENST00000511442.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr1_-_152297679 | 0.03 |
ENST00000368799.1
|
FLG
|
filaggrin |
| chr2_-_220042825 | 0.03 |
ENST00000409789.1
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr17_-_1418972 | 0.03 |
ENST00000571274.1
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr21_-_16374688 | 0.03 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr5_-_13944652 | 0.03 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr6_+_32121908 | 0.03 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr17_+_74261277 | 0.03 |
ENST00000327490.6
|
UBALD2
|
UBA-like domain containing 2 |
| chr8_+_120885949 | 0.03 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr11_-_72504637 | 0.03 |
ENST00000536377.1
ENST00000359373.5 |
STARD10
ARAP1
|
StAR-related lipid transfer (START) domain containing 10 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr12_+_52203789 | 0.03 |
ENST00000599343.1
|
AC068987.1
|
HCG1997999; cDNA FLJ33996 fis, clone DFNES2008881 |
| chr1_+_174933899 | 0.03 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr17_-_6947225 | 0.03 |
ENST00000574600.1
ENST00000308009.1 ENST00000447225.1 |
SLC16A11
|
solute carrier family 16, member 11 |
| chr9_-_21482312 | 0.03 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr15_-_34610962 | 0.03 |
ENST00000290209.5
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr16_-_4665023 | 0.03 |
ENST00000591897.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr3_-_42306248 | 0.03 |
ENST00000334681.5
|
CCK
|
cholecystokinin |
| chr12_-_772901 | 0.03 |
ENST00000305108.4
|
NINJ2
|
ninjurin 2 |
| chr9_+_131904233 | 0.03 |
ENST00000432651.1
ENST00000435132.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr1_+_61542922 | 0.03 |
ENST00000407417.3
|
NFIA
|
nuclear factor I/A |
| chr19_-_56047661 | 0.03 |
ENST00000344158.3
|
SBK2
|
SH3 domain binding kinase family, member 2 |
| chr9_+_6413317 | 0.03 |
ENST00000276893.5
ENST00000381373.3 |
UHRF2
|
ubiquitin-like with PHD and ring finger domains 2, E3 ubiquitin protein ligase |
| chr5_-_146833803 | 0.03 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr6_+_64345698 | 0.03 |
ENST00000506783.1
ENST00000481385.2 ENST00000515594.1 ENST00000494284.2 ENST00000262043.3 |
PHF3
|
PHD finger protein 3 |
| chr3_-_141868293 | 0.03 |
ENST00000317104.7
ENST00000494358.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr3_+_107241783 | 0.03 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr1_+_164528437 | 0.03 |
ENST00000485769.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr10_-_90751038 | 0.03 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr9_-_98269481 | 0.03 |
ENST00000418258.1
ENST00000553011.1 ENST00000551845.1 |
PTCH1
|
patched 1 |
| chr19_+_12902289 | 0.03 |
ENST00000302754.4
|
JUNB
|
jun B proto-oncogene |
| chrX_+_119005399 | 0.03 |
ENST00000371437.4
|
NDUFA1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1, 7.5kDa |
| chr7_+_134576317 | 0.03 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr14_-_24584138 | 0.03 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr14_+_50234827 | 0.03 |
ENST00000554589.1
ENST00000557247.1 |
KLHDC2
|
kelch domain containing 2 |
| chr7_+_91570165 | 0.03 |
ENST00000356239.3
ENST00000359028.2 ENST00000358100.2 |
AKAP9
|
A kinase (PRKA) anchor protein 9 |
| chr5_-_148758839 | 0.03 |
ENST00000261796.3
|
IL17B
|
interleukin 17B |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXD1_FOXO1_FOXO6_FOXG1_FOXP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.0 | 0.1 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.1 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:1905072 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.0 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.0 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.2 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.0 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.0 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.0 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.0 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.0 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.0 | GO:0036457 | keratohyalin granule(GO:0036457) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0004974 | leukotriene B4 receptor activity(GO:0001632) leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |