Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
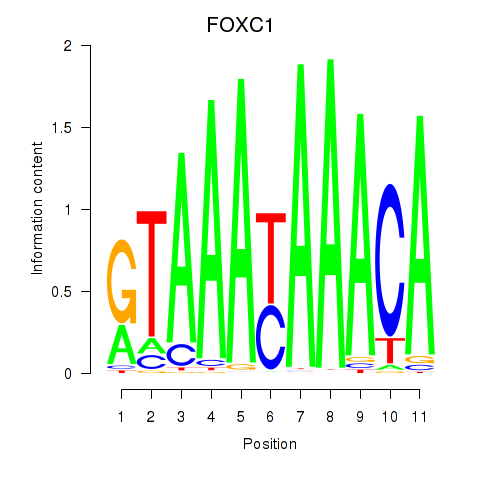
Results for FOXC1
Z-value: 0.38
Transcription factors associated with FOXC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXC1
|
ENSG00000054598.5 | forkhead box C1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXC1 | hg19_v2_chr6_+_1610681_1610681 | 0.98 | 1.5e-02 | Click! |
Activity profile of FOXC1 motif
Sorted Z-values of FOXC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_53303123 | 0.27 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr8_-_145754428 | 0.21 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr4_-_105416039 | 0.19 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr16_+_2570340 | 0.18 |
ENST00000568263.1
ENST00000293971.6 ENST00000302956.4 ENST00000413459.3 ENST00000566706.1 ENST00000569879.1 |
AMDHD2
|
amidohydrolase domain containing 2 |
| chr2_+_66918558 | 0.18 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr12_-_49453557 | 0.18 |
ENST00000547610.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr3_+_72201910 | 0.16 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr19_-_47734448 | 0.15 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr2_-_72375167 | 0.15 |
ENST00000001146.2
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr2_+_162272605 | 0.15 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr3_-_178865747 | 0.14 |
ENST00000435560.1
|
RP11-360P21.2
|
RP11-360P21.2 |
| chr2_-_25451065 | 0.14 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr20_+_43990576 | 0.13 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr2_+_166095898 | 0.13 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr10_+_71561630 | 0.13 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr3_-_134092561 | 0.12 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr22_-_50970506 | 0.12 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr10_+_71561649 | 0.12 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr17_+_72426891 | 0.12 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr17_+_79953310 | 0.11 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr5_+_95385819 | 0.11 |
ENST00000507997.1
|
RP11-254I22.1
|
RP11-254I22.1 |
| chr2_-_72374948 | 0.11 |
ENST00000546307.1
ENST00000474509.1 |
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr14_-_54418598 | 0.11 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr22_-_50970919 | 0.11 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr2_+_173724771 | 0.10 |
ENST00000538974.1
ENST00000540783.1 |
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_+_173686303 | 0.10 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr7_+_28452130 | 0.10 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr2_+_85981008 | 0.10 |
ENST00000306279.3
|
ATOH8
|
atonal homolog 8 (Drosophila) |
| chr16_-_71323617 | 0.10 |
ENST00000563876.1
|
CMTR2
|
cap methyltransferase 2 |
| chr19_-_40919271 | 0.10 |
ENST00000291825.7
ENST00000324001.7 |
PRX
|
periaxin |
| chr15_-_76352069 | 0.10 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr3_-_11623804 | 0.09 |
ENST00000451674.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr16_+_2570431 | 0.09 |
ENST00000563556.1
|
AMDHD2
|
amidohydrolase domain containing 2 |
| chr16_-_4817129 | 0.09 |
ENST00000545009.1
ENST00000219478.6 |
ZNF500
|
zinc finger protein 500 |
| chr22_-_39268308 | 0.09 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr8_-_108510224 | 0.09 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr7_-_37026108 | 0.09 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr11_-_790060 | 0.09 |
ENST00000330106.4
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr1_-_228135599 | 0.08 |
ENST00000272164.5
|
WNT9A
|
wingless-type MMTV integration site family, member 9A |
| chr5_+_150040403 | 0.08 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr3_-_127455200 | 0.08 |
ENST00000398101.3
|
MGLL
|
monoglyceride lipase |
| chr2_+_105050794 | 0.08 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr20_-_62587735 | 0.08 |
ENST00000354216.6
ENST00000369892.3 ENST00000358711.3 |
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr9_-_139258235 | 0.08 |
ENST00000371738.3
|
DNLZ
|
DNL-type zinc finger |
| chr11_-_46638720 | 0.08 |
ENST00000326737.3
|
HARBI1
|
harbinger transposase derived 1 |
| chr12_+_43086018 | 0.08 |
ENST00000550177.1
|
RP11-25I15.3
|
RP11-25I15.3 |
| chr11_+_46638805 | 0.08 |
ENST00000434074.1
ENST00000312040.4 ENST00000451945.1 |
ATG13
|
autophagy related 13 |
| chr11_-_104480019 | 0.08 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr5_-_55529115 | 0.08 |
ENST00000513241.2
ENST00000341048.4 |
ANKRD55
|
ankyrin repeat domain 55 |
| chr1_+_12524965 | 0.08 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr4_+_124571409 | 0.07 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr14_-_61191049 | 0.07 |
ENST00000556952.3
|
SIX4
|
SIX homeobox 4 |
| chr5_+_112312416 | 0.07 |
ENST00000389063.2
|
DCP2
|
decapping mRNA 2 |
| chr1_-_100231349 | 0.07 |
ENST00000287474.5
ENST00000414213.1 |
FRRS1
|
ferric-chelate reductase 1 |
| chr19_+_36024310 | 0.07 |
ENST00000222286.4
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr17_+_8924837 | 0.07 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr19_+_782755 | 0.07 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr19_+_42806250 | 0.07 |
ENST00000598490.1
ENST00000341747.3 |
PRR19
|
proline rich 19 |
| chr22_-_50970566 | 0.07 |
ENST00000405135.1
ENST00000401779.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr20_-_62582475 | 0.07 |
ENST00000369908.5
|
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr16_-_46649905 | 0.07 |
ENST00000569702.1
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr2_-_224467002 | 0.07 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr11_+_110225855 | 0.07 |
ENST00000526605.1
ENST00000526703.1 |
RP11-347E10.1
|
RP11-347E10.1 |
| chr14_+_23340822 | 0.07 |
ENST00000359591.4
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr19_+_17516531 | 0.06 |
ENST00000528911.1
ENST00000528604.1 ENST00000595892.1 ENST00000500836.2 ENST00000598546.1 ENST00000600369.1 ENST00000598356.1 ENST00000594426.1 |
MVB12A
CTD-2521M24.9
|
multivesicular body subunit 12A CTD-2521M24.9 |
| chr6_-_42016385 | 0.06 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr7_+_139528952 | 0.06 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr16_-_740419 | 0.06 |
ENST00000248142.6
|
WDR24
|
WD repeat domain 24 |
| chr11_-_46615498 | 0.06 |
ENST00000533727.1
ENST00000534300.1 ENST00000528950.1 ENST00000526606.1 |
AMBRA1
|
autophagy/beclin-1 regulator 1 |
| chr7_-_72972319 | 0.06 |
ENST00000223368.2
|
BCL7B
|
B-cell CLL/lymphoma 7B |
| chr5_-_115872124 | 0.06 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr4_-_102268628 | 0.06 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr10_-_118032979 | 0.06 |
ENST00000355422.6
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr22_-_43398442 | 0.06 |
ENST00000422336.1
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr17_+_28268623 | 0.06 |
ENST00000394835.3
ENST00000320856.5 ENST00000394832.2 ENST00000378738.3 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr11_-_126870655 | 0.06 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr2_-_220042825 | 0.06 |
ENST00000409789.1
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr1_+_223101757 | 0.06 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr12_-_92539614 | 0.06 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr11_-_8739566 | 0.06 |
ENST00000533020.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr1_+_220701456 | 0.06 |
ENST00000366918.4
ENST00000402574.1 |
MARK1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr9_-_139258159 | 0.06 |
ENST00000371739.3
|
DNLZ
|
DNL-type zinc finger |
| chr7_+_139529040 | 0.06 |
ENST00000455353.1
ENST00000458722.1 ENST00000411653.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr10_+_71562180 | 0.06 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr6_+_84563295 | 0.06 |
ENST00000369687.1
|
RIPPLY2
|
ripply transcriptional repressor 2 |
| chr1_-_153931052 | 0.05 |
ENST00000368630.3
ENST00000368633.1 |
CRTC2
|
CREB regulated transcription coactivator 2 |
| chr12_-_116714564 | 0.05 |
ENST00000548743.1
|
MED13L
|
mediator complex subunit 13-like |
| chr4_-_40477766 | 0.05 |
ENST00000507180.1
|
RBM47
|
RNA binding motif protein 47 |
| chr19_+_17516909 | 0.05 |
ENST00000601007.1
ENST00000594913.1 ENST00000599975.1 ENST00000600514.1 |
CTD-2521M24.9
MVB12A
|
CTD-2521M24.9 multivesicular body subunit 12A |
| chr11_+_46639071 | 0.05 |
ENST00000580238.1
ENST00000581416.1 ENST00000529655.1 ENST00000533325.1 ENST00000581438.1 ENST00000583249.1 ENST00000530500.1 ENST00000526508.1 ENST00000578626.1 ENST00000577256.1 ENST00000524625.1 ENST00000582547.1 ENST00000359513.4 ENST00000528494.1 |
ATG13
|
autophagy related 13 |
| chr14_+_51026844 | 0.05 |
ENST00000554886.1
|
ATL1
|
atlastin GTPase 1 |
| chr6_+_32121908 | 0.05 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr3_-_168865522 | 0.05 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr15_+_63889577 | 0.05 |
ENST00000534939.1
ENST00000539570.3 |
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chr12_+_122516626 | 0.05 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chr15_+_74509530 | 0.05 |
ENST00000321288.5
|
CCDC33
|
coiled-coil domain containing 33 |
| chr14_+_76776957 | 0.05 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr6_+_32121789 | 0.05 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr3_+_46448648 | 0.05 |
ENST00000399036.3
|
CCRL2
|
chemokine (C-C motif) receptor-like 2 |
| chr1_-_28384598 | 0.04 |
ENST00000373864.1
|
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chr5_-_124082279 | 0.04 |
ENST00000513986.1
|
ZNF608
|
zinc finger protein 608 |
| chr1_+_150122034 | 0.04 |
ENST00000025469.6
ENST00000369124.4 |
PLEKHO1
|
pleckstrin homology domain containing, family O member 1 |
| chr19_+_3880581 | 0.04 |
ENST00000450849.2
ENST00000301260.6 ENST00000398448.3 |
ATCAY
|
ataxia, cerebellar, Cayman type |
| chr21_+_17792672 | 0.04 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr8_-_134309823 | 0.04 |
ENST00000414097.2
|
NDRG1
|
N-myc downstream regulated 1 |
| chr19_-_36523529 | 0.04 |
ENST00000593074.1
|
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr3_-_49066811 | 0.04 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr1_+_207277590 | 0.04 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr10_+_71561704 | 0.04 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chrX_+_22050546 | 0.04 |
ENST00000379374.4
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr10_+_54074033 | 0.04 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr1_+_18958008 | 0.04 |
ENST00000420770.2
ENST00000400661.3 |
PAX7
|
paired box 7 |
| chr8_-_134309335 | 0.04 |
ENST00000522890.1
ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1
|
N-myc downstream regulated 1 |
| chr4_-_99578789 | 0.04 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chr11_-_66112555 | 0.04 |
ENST00000425825.2
ENST00000359957.3 |
BRMS1
|
breast cancer metastasis suppressor 1 |
| chr7_-_99698338 | 0.04 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr1_+_155099927 | 0.04 |
ENST00000368407.3
|
EFNA1
|
ephrin-A1 |
| chr6_-_152639479 | 0.04 |
ENST00000356820.4
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr4_-_153303658 | 0.04 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr16_+_8715574 | 0.04 |
ENST00000561758.1
|
METTL22
|
methyltransferase like 22 |
| chr16_+_30064411 | 0.04 |
ENST00000338110.5
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr3_-_57326704 | 0.04 |
ENST00000487349.1
ENST00000389601.3 |
ASB14
|
ankyrin repeat and SOCS box containing 14 |
| chr16_+_30064444 | 0.04 |
ENST00000395248.1
ENST00000566897.1 ENST00000568435.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr18_+_60382672 | 0.04 |
ENST00000400316.4
ENST00000262719.5 |
PHLPP1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr9_-_4299874 | 0.04 |
ENST00000381971.3
ENST00000477901.1 |
GLIS3
|
GLIS family zinc finger 3 |
| chr3_-_139195350 | 0.04 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr4_+_15376165 | 0.04 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr16_-_740354 | 0.04 |
ENST00000293883.4
|
WDR24
|
WD repeat domain 24 |
| chr1_-_149982624 | 0.04 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chr3_+_69812877 | 0.04 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr1_-_246729544 | 0.04 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr2_-_190044480 | 0.04 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr17_+_17685422 | 0.04 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr12_+_131438443 | 0.04 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr20_+_44509857 | 0.03 |
ENST00000372523.1
ENST00000372520.1 |
ZSWIM1
|
zinc finger, SWIM-type containing 1 |
| chr11_-_6440283 | 0.03 |
ENST00000299402.6
ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr1_+_43855560 | 0.03 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr10_-_118031778 | 0.03 |
ENST00000369236.1
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr2_+_173792893 | 0.03 |
ENST00000535187.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr1_-_94079648 | 0.03 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr20_-_22566089 | 0.03 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr18_+_19749386 | 0.03 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr1_+_73771844 | 0.03 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr19_-_4831701 | 0.03 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
| chr2_-_208030647 | 0.03 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr3_-_48481434 | 0.03 |
ENST00000395694.2
ENST00000447018.1 ENST00000442740.1 |
CCDC51
|
coiled-coil domain containing 51 |
| chr16_+_56969284 | 0.03 |
ENST00000568358.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr17_-_48546232 | 0.03 |
ENST00000258969.4
|
CHAD
|
chondroadherin |
| chr6_-_10747802 | 0.03 |
ENST00000606522.1
ENST00000606652.1 |
RP11-421M1.8
|
RP11-421M1.8 |
| chr3_-_24207039 | 0.03 |
ENST00000280696.5
|
THRB
|
thyroid hormone receptor, beta |
| chr15_-_72563585 | 0.03 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr5_+_112312399 | 0.03 |
ENST00000515408.1
ENST00000513585.1 |
DCP2
|
decapping mRNA 2 |
| chr12_+_10365082 | 0.03 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr6_+_15401075 | 0.03 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr4_+_86396321 | 0.03 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr10_+_114710425 | 0.03 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr3_-_48481518 | 0.03 |
ENST00000412398.2
ENST00000395696.1 |
CCDC51
|
coiled-coil domain containing 51 |
| chr14_+_38033252 | 0.03 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chr3_+_46449049 | 0.03 |
ENST00000357392.4
ENST00000400880.3 ENST00000433848.1 |
CCRL2
|
chemokine (C-C motif) receptor-like 2 |
| chr5_-_115872142 | 0.03 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr10_-_4285923 | 0.03 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr1_-_151431647 | 0.03 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr13_+_36050881 | 0.03 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr18_-_53070913 | 0.03 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr1_+_56046710 | 0.03 |
ENST00000422374.1
|
RP11-466L17.1
|
RP11-466L17.1 |
| chr15_+_96869165 | 0.03 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_+_58655461 | 0.03 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr15_-_55657428 | 0.03 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr1_-_43855444 | 0.02 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr17_-_57229155 | 0.02 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr2_+_181845532 | 0.02 |
ENST00000602475.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chrX_+_9431324 | 0.02 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr12_+_59989791 | 0.02 |
ENST00000552432.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr5_+_40841276 | 0.02 |
ENST00000254691.5
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr17_+_40925454 | 0.02 |
ENST00000253794.2
ENST00000590339.1 ENST00000589520.1 |
VPS25
|
vacuolar protein sorting 25 homolog (S. cerevisiae) |
| chr11_-_6440624 | 0.02 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr15_+_63889552 | 0.02 |
ENST00000360587.2
|
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chr2_+_172309634 | 0.02 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr1_-_57045228 | 0.02 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr15_+_67418047 | 0.02 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr6_-_152489484 | 0.02 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr12_-_49333446 | 0.02 |
ENST00000537495.1
|
AC073610.5
|
Uncharacterized protein |
| chr6_-_32122106 | 0.02 |
ENST00000428778.1
|
PRRT1
|
proline-rich transmembrane protein 1 |
| chr8_+_31497271 | 0.02 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr2_-_224467093 | 0.02 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr2_+_28974668 | 0.02 |
ENST00000296122.6
ENST00000395366.2 |
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr14_-_20774092 | 0.02 |
ENST00000423949.2
ENST00000553828.1 ENST00000258821.3 |
TTC5
|
tetratricopeptide repeat domain 5 |
| chr2_+_181845843 | 0.02 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr19_-_13900972 | 0.02 |
ENST00000397557.1
|
AC008686.1
|
Uncharacterized protein |
| chr1_-_94312706 | 0.02 |
ENST00000370244.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr12_-_53994805 | 0.02 |
ENST00000328463.7
|
ATF7
|
activating transcription factor 7 |
| chr3_-_129279894 | 0.02 |
ENST00000506979.1
|
PLXND1
|
plexin D1 |
| chr12_-_122018346 | 0.02 |
ENST00000377069.4
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr10_+_69869237 | 0.02 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr12_-_123756781 | 0.02 |
ENST00000544658.1
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr19_+_29493486 | 0.02 |
ENST00000589821.1
|
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr3_-_187454281 | 0.02 |
ENST00000232014.4
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr6_+_34204642 | 0.02 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr14_-_24020858 | 0.02 |
ENST00000419474.3
|
ZFHX2
|
zinc finger homeobox 2 |
| chr7_+_136553370 | 0.02 |
ENST00000445907.2
|
CHRM2
|
cholinergic receptor, muscarinic 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:2001035 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.2 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.1 | GO:0072096 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.0 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.0 | GO:0090381 | negative regulation of mesodermal cell fate specification(GO:0042662) motor learning(GO:0061743) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.0 | 0.1 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) ceramide binding(GO:0097001) |