Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
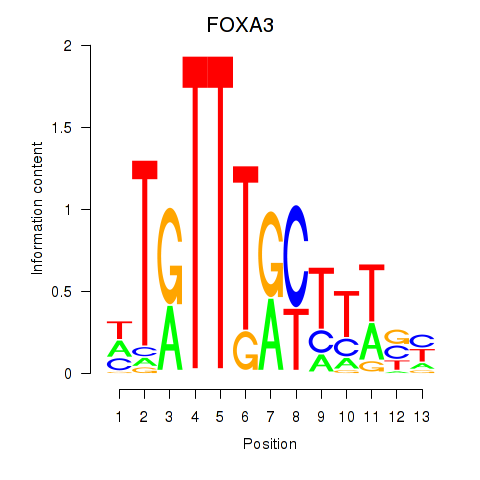
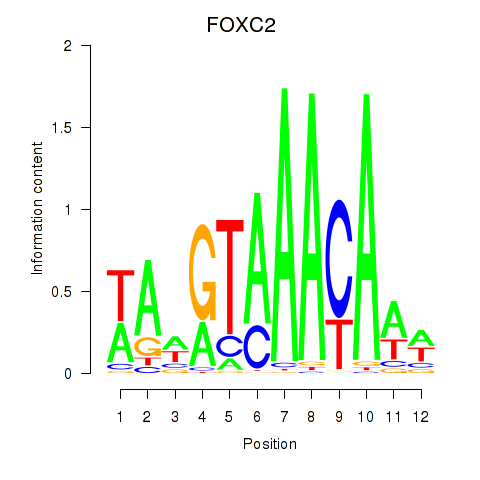
Results for FOXA3_FOXC2
Z-value: 0.38
Transcription factors associated with FOXA3_FOXC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA3
|
ENSG00000170608.2 | forkhead box A3 |
|
FOXC2
|
ENSG00000176692.4 | forkhead box C2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA3 | hg19_v2_chr19_+_46367518_46367567 | -0.61 | 3.9e-01 | Click! |
| FOXC2 | hg19_v2_chr16_+_86600857_86600921 | 0.10 | 9.0e-01 | Click! |
Activity profile of FOXA3_FOXC2 motif
Sorted Z-values of FOXA3_FOXC2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_106415457 | 0.16 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr1_-_207095324 | 0.12 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr2_+_179318295 | 0.12 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr2_+_58655461 | 0.11 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr1_-_149859466 | 0.11 |
ENST00000331128.3
|
HIST2H2AB
|
histone cluster 2, H2ab |
| chr12_-_12714006 | 0.11 |
ENST00000541207.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr1_-_1356628 | 0.10 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr1_-_1356719 | 0.10 |
ENST00000520296.1
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr12_-_12714025 | 0.10 |
ENST00000539940.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr4_+_76871883 | 0.10 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr2_+_109223595 | 0.09 |
ENST00000410093.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr2_-_208030295 | 0.09 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr10_+_124739964 | 0.09 |
ENST00000406217.2
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr5_+_95998746 | 0.08 |
ENST00000508608.1
|
CAST
|
calpastatin |
| chr16_-_87970122 | 0.08 |
ENST00000309893.2
|
CA5A
|
carbonic anhydrase VA, mitochondrial |
| chr2_-_87248975 | 0.08 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr17_-_19290483 | 0.08 |
ENST00000395592.2
ENST00000299610.4 |
MFAP4
|
microfibrillar-associated protein 4 |
| chr5_-_137674000 | 0.07 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr11_+_65851443 | 0.07 |
ENST00000533756.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr12_+_96588279 | 0.07 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr11_+_8040739 | 0.07 |
ENST00000534099.1
|
TUB
|
tubby bipartite transcription factor |
| chr10_-_128210005 | 0.07 |
ENST00000284694.7
ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr2_+_196521903 | 0.06 |
ENST00000541054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr7_+_151791074 | 0.06 |
ENST00000447796.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr9_-_20382446 | 0.06 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr8_+_142264664 | 0.06 |
ENST00000518520.1
|
RP11-10J21.3
|
Uncharacterized protein |
| chr6_-_13621126 | 0.06 |
ENST00000600057.1
|
AL441883.1
|
Uncharacterized protein |
| chr2_+_196521845 | 0.06 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr7_+_151791037 | 0.06 |
ENST00000419245.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr2_-_175711924 | 0.06 |
ENST00000444573.1
|
CHN1
|
chimerin 1 |
| chr6_+_37897735 | 0.06 |
ENST00000373389.5
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr3_-_193096600 | 0.06 |
ENST00000446087.1
ENST00000342358.4 |
ATP13A5
|
ATPase type 13A5 |
| chr17_-_36358166 | 0.05 |
ENST00000537432.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr12_+_59989918 | 0.05 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_+_152486950 | 0.05 |
ENST00000368790.3
|
CRCT1
|
cysteine-rich C-terminal 1 |
| chr1_+_239882842 | 0.05 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr6_+_26251835 | 0.05 |
ENST00000356350.2
|
HIST1H2BH
|
histone cluster 1, H2bh |
| chr2_+_66918558 | 0.05 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr2_-_192711968 | 0.05 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr15_+_63188009 | 0.05 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr10_-_118032697 | 0.05 |
ENST00000439649.3
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr2_+_136343904 | 0.05 |
ENST00000436436.1
|
R3HDM1
|
R3H domain containing 1 |
| chr2_-_175711978 | 0.05 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr15_+_76016293 | 0.05 |
ENST00000332145.2
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1 |
| chr5_+_169011033 | 0.04 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr2_-_46844242 | 0.04 |
ENST00000281382.6
|
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr9_+_129097520 | 0.04 |
ENST00000436593.3
|
MVB12B
|
multivesicular body subunit 12B |
| chr18_-_52626622 | 0.04 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr5_+_74807886 | 0.04 |
ENST00000514296.1
|
POLK
|
polymerase (DNA directed) kappa |
| chr1_+_150488205 | 0.04 |
ENST00000416894.1
|
LINC00568
|
long intergenic non-protein coding RNA 568 |
| chr14_-_31926623 | 0.04 |
ENST00000356180.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr11_-_72852320 | 0.04 |
ENST00000422375.1
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr7_+_151038850 | 0.04 |
ENST00000355851.4
ENST00000566856.1 ENST00000470229.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr15_-_59041768 | 0.04 |
ENST00000402627.1
ENST00000396140.2 ENST00000559053.1 ENST00000561288.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr12_+_120740119 | 0.04 |
ENST00000536460.1
ENST00000202967.4 |
SIRT4
|
sirtuin 4 |
| chr17_-_41322332 | 0.04 |
ENST00000590740.1
|
RP11-242D8.1
|
RP11-242D8.1 |
| chr4_+_75174204 | 0.04 |
ENST00000332112.4
ENST00000514968.1 ENST00000503098.1 ENST00000502358.1 ENST00000509145.1 ENST00000505212.1 |
EPGN
|
epithelial mitogen |
| chr2_-_165424973 | 0.04 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr18_-_25616519 | 0.04 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr17_+_37030127 | 0.04 |
ENST00000419929.1
|
LASP1
|
LIM and SH3 protein 1 |
| chr2_+_169658928 | 0.04 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr3_+_159570722 | 0.04 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr7_+_134576317 | 0.03 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr13_+_95364963 | 0.03 |
ENST00000438290.2
|
SOX21-AS1
|
SOX21 antisense RNA 1 (head to head) |
| chr20_+_9146969 | 0.03 |
ENST00000416836.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr5_+_133859996 | 0.03 |
ENST00000512386.1
|
PHF15
|
jade family PHD finger 2 |
| chr2_+_169659121 | 0.03 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr17_-_77967433 | 0.03 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr4_-_170679024 | 0.03 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr2_-_175711133 | 0.03 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr8_+_104384616 | 0.03 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr12_-_48099754 | 0.03 |
ENST00000380650.4
|
RPAP3
|
RNA polymerase II associated protein 3 |
| chr16_-_15474904 | 0.03 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr12_-_15865844 | 0.03 |
ENST00000543612.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr1_+_246729815 | 0.03 |
ENST00000366511.1
|
CNST
|
consortin, connexin sorting protein |
| chr7_+_95115210 | 0.03 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr5_+_147774275 | 0.03 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr20_+_9049742 | 0.03 |
ENST00000437503.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr3_+_155588375 | 0.03 |
ENST00000295920.7
|
GMPS
|
guanine monphosphate synthase |
| chr3_+_136649311 | 0.03 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr3_-_196910721 | 0.03 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr3_-_107941230 | 0.03 |
ENST00000264538.3
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr3_-_105588231 | 0.03 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr12_-_21927736 | 0.03 |
ENST00000240662.2
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr1_-_150669604 | 0.03 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr2_+_175260514 | 0.03 |
ENST00000424069.1
ENST00000427038.1 |
SCRN3
|
secernin 3 |
| chr5_+_74807581 | 0.03 |
ENST00000241436.4
ENST00000352007.5 |
POLK
|
polymerase (DNA directed) kappa |
| chr11_-_72070206 | 0.03 |
ENST00000544382.1
|
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr18_+_21033239 | 0.03 |
ENST00000581585.1
ENST00000577501.1 |
RIOK3
|
RIO kinase 3 |
| chr3_-_196910477 | 0.03 |
ENST00000447466.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr3_+_69985734 | 0.03 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr12_-_53207842 | 0.03 |
ENST00000458244.2
|
KRT4
|
keratin 4 |
| chr13_-_99667960 | 0.03 |
ENST00000448493.2
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr12_+_27849378 | 0.03 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr1_+_104159999 | 0.03 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr15_+_80364901 | 0.03 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr3_+_132316081 | 0.03 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr3_+_57882024 | 0.03 |
ENST00000494088.1
|
SLMAP
|
sarcolemma associated protein |
| chr12_-_106480587 | 0.03 |
ENST00000548902.1
|
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr13_+_73629107 | 0.03 |
ENST00000539231.1
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr7_+_134464376 | 0.03 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr15_+_59664884 | 0.03 |
ENST00000558348.1
|
FAM81A
|
family with sequence similarity 81, member A |
| chr3_-_71294304 | 0.03 |
ENST00000498215.1
|
FOXP1
|
forkhead box P1 |
| chr14_+_20811766 | 0.03 |
ENST00000250416.5
ENST00000527915.1 |
PARP2
|
poly (ADP-ribose) polymerase 2 |
| chr16_-_3350614 | 0.03 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr2_-_65593784 | 0.03 |
ENST00000443619.2
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr2_+_68592305 | 0.03 |
ENST00000234313.7
|
PLEK
|
pleckstrin |
| chr7_+_26438187 | 0.03 |
ENST00000439120.1
ENST00000430548.1 ENST00000421862.1 ENST00000449537.1 ENST00000420774.1 ENST00000418758.2 |
AC004540.5
|
AC004540.5 |
| chr1_-_154150651 | 0.03 |
ENST00000302206.5
|
TPM3
|
tropomyosin 3 |
| chr7_-_117512264 | 0.02 |
ENST00000454375.1
|
CTTNBP2
|
cortactin binding protein 2 |
| chr16_-_48281305 | 0.02 |
ENST00000356608.2
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr8_-_80993010 | 0.02 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr1_+_246729724 | 0.02 |
ENST00000366513.4
ENST00000366512.3 |
CNST
|
consortin, connexin sorting protein |
| chr4_-_152149033 | 0.02 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr14_+_20215587 | 0.02 |
ENST00000331723.1
|
OR4Q3
|
olfactory receptor, family 4, subfamily Q, member 3 |
| chr22_+_21128167 | 0.02 |
ENST00000215727.5
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr8_+_21777159 | 0.02 |
ENST00000434536.1
ENST00000252512.9 |
XPO7
|
exportin 7 |
| chr17_-_37764128 | 0.02 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr2_+_175260451 | 0.02 |
ENST00000458563.1
ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3
|
secernin 3 |
| chr10_-_126849588 | 0.02 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
| chr1_+_61547405 | 0.02 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr1_-_169555709 | 0.02 |
ENST00000546081.1
|
F5
|
coagulation factor V (proaccelerin, labile factor) |
| chr1_-_43855444 | 0.02 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr1_-_149459549 | 0.02 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr8_+_19171128 | 0.02 |
ENST00000265807.3
|
SH2D4A
|
SH2 domain containing 4A |
| chr10_-_118032979 | 0.02 |
ENST00000355422.6
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr4_+_86396321 | 0.02 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr8_+_97597148 | 0.02 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr5_+_61708582 | 0.02 |
ENST00000325324.6
|
IPO11
|
importin 11 |
| chr11_-_116708302 | 0.02 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr12_-_71551652 | 0.02 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr17_-_39684550 | 0.02 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chr14_+_58711539 | 0.02 |
ENST00000216455.4
ENST00000412908.2 ENST00000557508.1 |
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3 |
| chr15_+_69854027 | 0.02 |
ENST00000498938.2
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr1_+_114471972 | 0.02 |
ENST00000369559.4
ENST00000369554.2 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr3_-_139195350 | 0.02 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr5_+_140710061 | 0.02 |
ENST00000517417.1
ENST00000378105.3 |
PCDHGA1
|
protocadherin gamma subfamily A, 1 |
| chr10_+_81466084 | 0.02 |
ENST00000342531.2
|
NUTM2B
|
NUT family member 2B |
| chr12_-_48099773 | 0.02 |
ENST00000432584.3
ENST00000005386.3 |
RPAP3
|
RNA polymerase II associated protein 3 |
| chr14_-_24664540 | 0.02 |
ENST00000530563.1
ENST00000528895.1 ENST00000528669.1 ENST00000532632.1 |
TM9SF1
|
transmembrane 9 superfamily member 1 |
| chr10_-_21186144 | 0.02 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr14_+_20811722 | 0.02 |
ENST00000429687.3
|
PARP2
|
poly (ADP-ribose) polymerase 2 |
| chr3_+_148447887 | 0.02 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr4_+_71588372 | 0.02 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr14_+_56127989 | 0.02 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr21_-_36259445 | 0.02 |
ENST00000399240.1
|
RUNX1
|
runt-related transcription factor 1 |
| chr5_+_32531893 | 0.02 |
ENST00000512913.1
|
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr7_-_107643674 | 0.02 |
ENST00000222399.6
|
LAMB1
|
laminin, beta 1 |
| chr4_+_169013666 | 0.02 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr2_-_21266935 | 0.02 |
ENST00000233242.1
|
APOB
|
apolipoprotein B |
| chr9_+_116343192 | 0.02 |
ENST00000471324.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr1_-_177939041 | 0.02 |
ENST00000308284.6
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr2_+_171640291 | 0.02 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chrX_-_47863348 | 0.02 |
ENST00000376943.3
ENST00000396965.1 ENST00000305127.6 |
ZNF182
|
zinc finger protein 182 |
| chr7_+_134430212 | 0.02 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr6_-_131211534 | 0.02 |
ENST00000456097.2
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr4_-_21950356 | 0.02 |
ENST00000447367.2
ENST00000382152.2 |
KCNIP4
|
Kv channel interacting protein 4 |
| chr15_+_71185148 | 0.02 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr8_+_101349823 | 0.02 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr4_+_79567362 | 0.02 |
ENST00000512322.1
|
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr7_+_77469439 | 0.02 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr16_+_4666475 | 0.02 |
ENST00000591895.1
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr20_+_9049682 | 0.02 |
ENST00000334005.3
ENST00000378473.3 |
PLCB4
|
phospholipase C, beta 4 |
| chr10_-_31288398 | 0.02 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr14_+_56127960 | 0.02 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr3_+_130569592 | 0.02 |
ENST00000533801.2
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr2_-_161056762 | 0.02 |
ENST00000428609.2
ENST00000409967.2 |
ITGB6
|
integrin, beta 6 |
| chr6_+_21666633 | 0.02 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr3_-_116163830 | 0.02 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr4_+_148653206 | 0.02 |
ENST00000336498.3
|
ARHGAP10
|
Rho GTPase activating protein 10 |
| chr3_+_141106643 | 0.02 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr2_-_21266816 | 0.02 |
ENST00000399256.4
|
APOB
|
apolipoprotein B |
| chr15_+_59665194 | 0.02 |
ENST00000560394.1
|
FAM81A
|
family with sequence similarity 81, member A |
| chr5_-_58571935 | 0.02 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr19_+_15160130 | 0.02 |
ENST00000427043.3
|
CASP14
|
caspase 14, apoptosis-related cysteine peptidase |
| chr1_+_114471809 | 0.02 |
ENST00000426820.2
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr2_+_136343820 | 0.02 |
ENST00000410054.1
|
R3HDM1
|
R3H domain containing 1 |
| chr4_+_146403912 | 0.02 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr15_+_69857515 | 0.02 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr17_-_6947225 | 0.02 |
ENST00000574600.1
ENST00000308009.1 ENST00000447225.1 |
SLC16A11
|
solute carrier family 16, member 11 |
| chr1_+_12806141 | 0.02 |
ENST00000288048.5
|
C1orf158
|
chromosome 1 open reading frame 158 |
| chr2_+_28974489 | 0.02 |
ENST00000455580.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr3_+_142315225 | 0.02 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr7_+_134464414 | 0.02 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr5_+_95998714 | 0.02 |
ENST00000506811.1
ENST00000514055.1 |
CAST
|
calpastatin |
| chr3_+_142315294 | 0.02 |
ENST00000464320.1
|
PLS1
|
plastin 1 |
| chr5_+_65222500 | 0.02 |
ENST00000511297.1
ENST00000506030.1 |
ERBB2IP
|
erbb2 interacting protein |
| chr2_+_88047606 | 0.02 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr5_+_156712372 | 0.02 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr18_+_21032781 | 0.02 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr10_+_95848824 | 0.02 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr7_+_112063192 | 0.02 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr7_+_134576151 | 0.02 |
ENST00000393118.2
|
CALD1
|
caldesmon 1 |
| chr6_-_112575758 | 0.02 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr11_-_102401469 | 0.02 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr5_+_61874562 | 0.02 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr6_-_24666819 | 0.02 |
ENST00000341060.3
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr1_+_179851893 | 0.02 |
ENST00000531630.2
|
TOR1AIP1
|
torsin A interacting protein 1 |
| chr6_+_10528560 | 0.02 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr15_+_49462397 | 0.02 |
ENST00000396509.2
|
GALK2
|
galactokinase 2 |
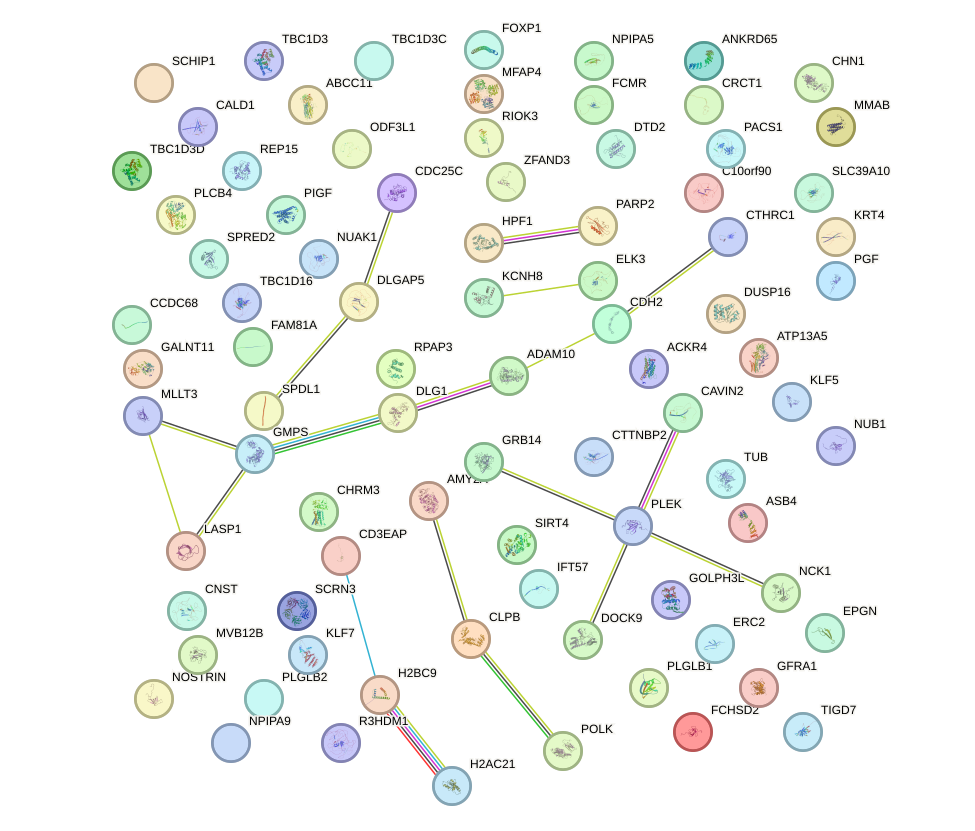
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA3_FOXC2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.1 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.0 | GO:1903217 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.1 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.0 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.0 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0034371 | chylomicron remodeling(GO:0034371) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.0 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.0 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.0 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |