Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
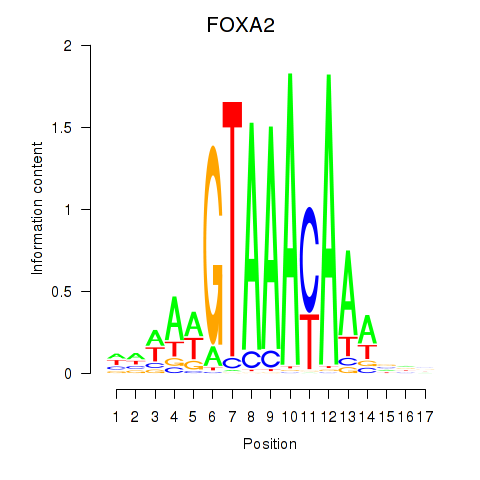
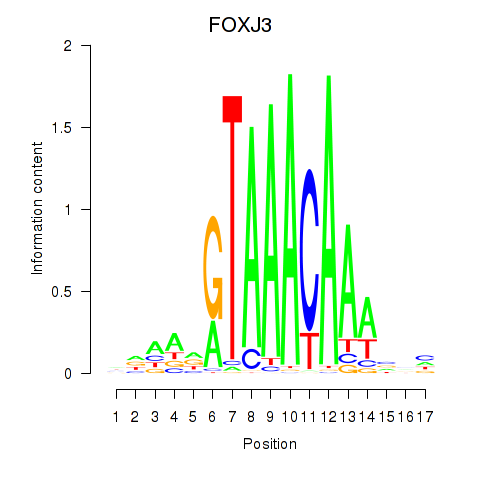
Results for FOXA2_FOXJ3
Z-value: 0.97
Transcription factors associated with FOXA2_FOXJ3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA2
|
ENSG00000125798.10 | forkhead box A2 |
|
FOXJ3
|
ENSG00000198815.4 | forkhead box J3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA2 | hg19_v2_chr20_-_22565101_22565223 | -0.74 | 2.6e-01 | Click! |
| FOXJ3 | hg19_v2_chr1_-_42800860_42800912 | 0.47 | 5.3e-01 | Click! |
Activity profile of FOXA2_FOXJ3 motif
Sorted Z-values of FOXA2_FOXJ3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_12714006 | 0.63 |
ENST00000541207.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr4_+_103790462 | 0.57 |
ENST00000503643.1
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chr10_+_111985837 | 0.53 |
ENST00000393134.1
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr1_+_161035655 | 0.53 |
ENST00000600454.1
|
AL591806.1
|
Uncharacterized protein |
| chr12_-_12714025 | 0.46 |
ENST00000539940.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr17_-_49021974 | 0.45 |
ENST00000501718.2
|
RP11-700H6.1
|
RP11-700H6.1 |
| chr1_+_246729815 | 0.42 |
ENST00000366511.1
|
CNST
|
consortin, connexin sorting protein |
| chr2_+_109223595 | 0.42 |
ENST00000410093.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr8_-_80993010 | 0.41 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr3_+_159570722 | 0.41 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr10_-_14050522 | 0.38 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr5_-_57854070 | 0.33 |
ENST00000504333.1
|
CTD-2117L12.1
|
Uncharacterized protein |
| chr5_-_159846066 | 0.32 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr1_-_238108575 | 0.32 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr5_+_159436120 | 0.29 |
ENST00000522793.1
ENST00000231238.5 |
TTC1
|
tetratricopeptide repeat domain 1 |
| chr5_-_137674000 | 0.29 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr2_-_87248975 | 0.29 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr5_+_169011033 | 0.29 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr7_+_151038850 | 0.29 |
ENST00000355851.4
ENST00000566856.1 ENST00000470229.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr11_+_65851443 | 0.28 |
ENST00000533756.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr11_-_85780853 | 0.28 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr16_+_10479906 | 0.28 |
ENST00000562527.1
ENST00000396560.2 ENST00000396559.1 ENST00000562102.1 ENST00000543967.1 ENST00000569939.1 ENST00000569900.1 |
ATF7IP2
|
activating transcription factor 7 interacting protein 2 |
| chr11_+_34643600 | 0.28 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr1_-_207095324 | 0.28 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr2_+_149402989 | 0.27 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr13_-_41240717 | 0.27 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr6_+_21666633 | 0.27 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr10_+_111985713 | 0.27 |
ENST00000239007.7
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr9_-_94877658 | 0.26 |
ENST00000262554.2
ENST00000337841.4 |
SPTLC1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr5_-_98262240 | 0.26 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr8_-_40755333 | 0.26 |
ENST00000297737.6
ENST00000315769.7 |
ZMAT4
|
zinc finger, matrin-type 4 |
| chr14_-_22938665 | 0.25 |
ENST00000535880.2
|
TRDV3
|
T cell receptor delta variable 3 |
| chr4_+_146403912 | 0.25 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr18_+_21033239 | 0.25 |
ENST00000581585.1
ENST00000577501.1 |
RIOK3
|
RIO kinase 3 |
| chr16_-_30265831 | 0.25 |
ENST00000568053.1
|
RP11-347C12.1
|
Putative NPIP-like protein LOC613037 |
| chr16_-_21442874 | 0.24 |
ENST00000534903.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr12_-_106480587 | 0.24 |
ENST00000548902.1
|
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr9_-_21305312 | 0.24 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr9_-_20382446 | 0.24 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr17_-_30228678 | 0.24 |
ENST00000261708.4
|
UTP6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chrX_-_47863348 | 0.24 |
ENST00000376943.3
ENST00000396965.1 ENST00000305127.6 |
ZNF182
|
zinc finger protein 182 |
| chr2_+_175260451 | 0.23 |
ENST00000458563.1
ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3
|
secernin 3 |
| chr2_+_109204909 | 0.23 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr16_-_80926457 | 0.23 |
ENST00000563626.1
ENST00000562231.1 |
RP11-314O13.1
|
RP11-314O13.1 |
| chr17_-_26662464 | 0.22 |
ENST00000579419.1
ENST00000585313.1 ENST00000395418.3 ENST00000578985.1 ENST00000577498.1 ENST00000585089.1 ENST00000357896.3 |
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr3_-_58523010 | 0.22 |
ENST00000459701.2
ENST00000302819.5 |
ACOX2
|
acyl-CoA oxidase 2, branched chain |
| chr2_+_191513959 | 0.22 |
ENST00000337386.5
ENST00000357215.5 |
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr2_-_37193606 | 0.22 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr12_-_68696652 | 0.22 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr4_+_26862313 | 0.22 |
ENST00000467087.1
ENST00000382009.3 ENST00000237364.5 |
STIM2
|
stromal interaction molecule 2 |
| chr3_+_132316081 | 0.21 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr15_+_57998923 | 0.21 |
ENST00000380557.4
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr10_+_124739964 | 0.20 |
ENST00000406217.2
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr15_+_35270552 | 0.20 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr1_+_92545862 | 0.20 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr19_-_52307357 | 0.20 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr6_-_52109335 | 0.20 |
ENST00000336123.4
|
IL17F
|
interleukin 17F |
| chr17_+_71229346 | 0.20 |
ENST00000535032.2
ENST00000582793.1 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr4_+_79567057 | 0.20 |
ENST00000503259.1
ENST00000507802.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr1_+_45140400 | 0.20 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr13_-_103346854 | 0.19 |
ENST00000267273.6
|
METTL21C
|
methyltransferase like 21C |
| chr3_+_171561127 | 0.19 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr4_+_165675197 | 0.19 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr6_-_13621126 | 0.19 |
ENST00000600057.1
|
AL441883.1
|
Uncharacterized protein |
| chr3_+_148447887 | 0.19 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr1_+_203765437 | 0.19 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr2_-_175260368 | 0.19 |
ENST00000342016.3
ENST00000362053.5 |
CIR1
|
corepressor interacting with RBPJ, 1 |
| chr3_-_185641681 | 0.19 |
ENST00000259043.7
|
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr14_+_97263641 | 0.19 |
ENST00000216639.3
|
VRK1
|
vaccinia related kinase 1 |
| chr15_+_79166065 | 0.19 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr1_+_95616933 | 0.19 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr16_+_53469525 | 0.19 |
ENST00000544405.2
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr12_-_21927736 | 0.19 |
ENST00000240662.2
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr4_-_21950356 | 0.18 |
ENST00000447367.2
ENST00000382152.2 |
KCNIP4
|
Kv channel interacting protein 4 |
| chr5_-_131132614 | 0.18 |
ENST00000307968.7
ENST00000307954.8 |
FNIP1
|
folliculin interacting protein 1 |
| chr6_+_123110465 | 0.18 |
ENST00000539041.1
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr17_+_22022437 | 0.18 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr4_+_117220016 | 0.18 |
ENST00000604093.1
|
MTRNR2L13
|
MT-RNR2-like 13 (pseudogene) |
| chr3_-_182703688 | 0.17 |
ENST00000466812.1
ENST00000487822.1 ENST00000460412.1 ENST00000469954.1 |
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr15_+_78857849 | 0.17 |
ENST00000299565.5
|
CHRNA5
|
cholinergic receptor, nicotinic, alpha 5 (neuronal) |
| chr15_-_59041768 | 0.17 |
ENST00000402627.1
ENST00000396140.2 ENST00000559053.1 ENST00000561288.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr6_+_122793058 | 0.17 |
ENST00000392491.2
|
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr14_-_102605983 | 0.17 |
ENST00000334701.7
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr8_+_103540983 | 0.17 |
ENST00000523572.1
|
KB-1980E6.3
|
Uncharacterized protein |
| chr17_-_64216748 | 0.17 |
ENST00000585162.1
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr2_+_3705785 | 0.17 |
ENST00000252505.3
|
ALLC
|
allantoicase |
| chr4_-_170679024 | 0.17 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr12_+_49297899 | 0.17 |
ENST00000552942.1
ENST00000320516.4 |
CCDC65
|
coiled-coil domain containing 65 |
| chr19_-_40732594 | 0.17 |
ENST00000430325.2
ENST00000433940.1 |
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr1_+_24018269 | 0.16 |
ENST00000374550.3
|
RPL11
|
ribosomal protein L11 |
| chr10_-_32217717 | 0.16 |
ENST00000396144.4
ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12
|
Rho GTPase activating protein 12 |
| chr9_+_70856397 | 0.16 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr12_-_122107549 | 0.16 |
ENST00000355329.3
|
MORN3
|
MORN repeat containing 3 |
| chr10_+_52724028 | 0.16 |
ENST00000561565.1
|
RP11-96B5.4
|
RP11-96B5.4 |
| chr20_+_33104199 | 0.16 |
ENST00000357156.2
ENST00000417166.2 ENST00000300469.9 ENST00000374846.3 |
DYNLRB1
|
dynein, light chain, roadblock-type 1 |
| chr7_+_26332645 | 0.16 |
ENST00000396376.1
|
SNX10
|
sorting nexin 10 |
| chr19_-_10311868 | 0.15 |
ENST00000588118.1
ENST00000586800.1 |
DNMT1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr11_-_5537920 | 0.15 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr1_+_33439268 | 0.15 |
ENST00000594612.1
|
FKSG48
|
FKSG48 |
| chr13_+_98086445 | 0.15 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr1_+_66796401 | 0.15 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr2_+_109204743 | 0.15 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr16_+_22518495 | 0.15 |
ENST00000541154.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr8_+_11666649 | 0.15 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr5_+_136070614 | 0.15 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr9_-_70490107 | 0.15 |
ENST00000377395.4
ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5
|
COBW domain containing 5 |
| chr1_+_150488205 | 0.14 |
ENST00000416894.1
|
LINC00568
|
long intergenic non-protein coding RNA 568 |
| chr13_+_73629107 | 0.14 |
ENST00000539231.1
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr16_-_86542455 | 0.14 |
ENST00000595886.1
ENST00000597578.1 ENST00000593604.1 |
FENDRR
|
FOXF1 adjacent non-coding developmental regulatory RNA |
| chr9_-_86432547 | 0.14 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr1_-_77685084 | 0.14 |
ENST00000370812.3
ENST00000359130.1 ENST00000445065.1 ENST00000370813.5 |
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr3_-_48956818 | 0.14 |
ENST00000408959.2
|
ARIH2OS
|
ariadne homolog 2 opposite strand |
| chr15_+_78857870 | 0.14 |
ENST00000559554.1
|
CHRNA5
|
cholinergic receptor, nicotinic, alpha 5 (neuronal) |
| chr3_-_107099454 | 0.14 |
ENST00000593837.1
ENST00000599431.1 |
RP11-446H18.5
|
RP11-446H18.5 |
| chr3_+_130650738 | 0.14 |
ENST00000504612.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr14_+_35747825 | 0.14 |
ENST00000540871.1
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr11_+_102217936 | 0.14 |
ENST00000532832.1
ENST00000530675.1 ENST00000533742.1 ENST00000227758.2 ENST00000532672.1 ENST00000531259.1 ENST00000527465.1 |
BIRC2
|
baculoviral IAP repeat containing 2 |
| chr1_-_24194771 | 0.14 |
ENST00000374479.3
|
FUCA1
|
fucosidase, alpha-L- 1, tissue |
| chr19_+_52839515 | 0.14 |
ENST00000403906.3
ENST00000601151.1 |
ZNF610
|
zinc finger protein 610 |
| chr19_-_39826639 | 0.14 |
ENST00000602185.1
ENST00000598034.1 ENST00000601387.1 ENST00000595636.1 ENST00000253054.8 ENST00000594700.1 ENST00000597595.1 |
GMFG
|
glia maturation factor, gamma |
| chr9_-_104198042 | 0.14 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate |
| chr8_+_21777159 | 0.13 |
ENST00000434536.1
ENST00000252512.9 |
XPO7
|
exportin 7 |
| chr6_-_131949200 | 0.13 |
ENST00000539158.1
ENST00000368058.1 |
MED23
|
mediator complex subunit 23 |
| chr6_-_131321863 | 0.13 |
ENST00000528282.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr2_+_114195268 | 0.13 |
ENST00000259199.4
ENST00000416503.2 ENST00000433343.2 |
CBWD2
|
COBW domain containing 2 |
| chr2_-_46844242 | 0.13 |
ENST00000281382.6
|
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr6_+_37012607 | 0.13 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr11_+_118398178 | 0.13 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chr1_-_114429997 | 0.13 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2-like 15 |
| chr19_+_41768561 | 0.13 |
ENST00000599719.1
ENST00000601309.1 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr8_-_93978216 | 0.13 |
ENST00000517751.1
ENST00000524107.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr7_-_139876734 | 0.13 |
ENST00000006967.5
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr14_-_58893832 | 0.13 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr10_+_70847852 | 0.13 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr5_+_179135246 | 0.13 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr18_+_21032781 | 0.13 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr14_+_104177607 | 0.12 |
ENST00000429169.1
|
AL049840.1
|
Uncharacterized protein; cDNA FLJ53535 |
| chr3_+_101292939 | 0.12 |
ENST00000265260.3
ENST00000469941.1 ENST00000296024.5 |
PCNP
|
PEST proteolytic signal containing nuclear protein |
| chr5_+_218356 | 0.12 |
ENST00000264932.6
ENST00000504309.1 ENST00000510361.1 |
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr1_+_246729724 | 0.12 |
ENST00000366513.4
ENST00000366512.3 |
CNST
|
consortin, connexin sorting protein |
| chr1_-_145610869 | 0.12 |
ENST00000334163.3
ENST00000369294.1 |
POLR3C
|
polymerase (RNA) III (DNA directed) polypeptide C (62kD) |
| chr4_-_111120334 | 0.12 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr6_-_80247140 | 0.12 |
ENST00000392959.1
ENST00000467898.3 |
LCA5
|
Leber congenital amaurosis 5 |
| chr1_-_28559502 | 0.12 |
ENST00000263697.4
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8 |
| chr7_+_128312346 | 0.12 |
ENST00000480462.1
ENST00000378704.3 ENST00000477515.1 |
FAM71F2
|
family with sequence similarity 71, member F2 |
| chr18_-_52626622 | 0.12 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr15_+_86098670 | 0.12 |
ENST00000558811.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr10_-_103578162 | 0.12 |
ENST00000361464.3
ENST00000357797.5 ENST00000370094.3 |
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr7_-_23571586 | 0.12 |
ENST00000538367.1
ENST00000392502.4 ENST00000297071.4 |
TRA2A
|
transformer 2 alpha homolog (Drosophila) |
| chr10_-_25010795 | 0.12 |
ENST00000416305.1
ENST00000376410.2 |
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr15_-_54051831 | 0.12 |
ENST00000557913.1
ENST00000360509.5 |
WDR72
|
WD repeat domain 72 |
| chr6_-_42185583 | 0.12 |
ENST00000053468.3
|
MRPS10
|
mitochondrial ribosomal protein S10 |
| chr5_+_70751442 | 0.12 |
ENST00000358731.4
ENST00000380675.2 |
BDP1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
| chr2_-_128615681 | 0.12 |
ENST00000409955.1
ENST00000272645.4 |
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr2_-_26251481 | 0.12 |
ENST00000599234.1
|
AC013449.1
|
Uncharacterized protein |
| chr8_+_48920960 | 0.12 |
ENST00000523111.2
ENST00000523432.1 ENST00000521346.1 ENST00000517630.1 |
UBE2V2
|
ubiquitin-conjugating enzyme E2 variant 2 |
| chr9_+_108463234 | 0.12 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr5_+_95998714 | 0.12 |
ENST00000506811.1
ENST00000514055.1 |
CAST
|
calpastatin |
| chr4_+_172734548 | 0.12 |
ENST00000506823.1
|
GALNTL6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr5_+_79703823 | 0.12 |
ENST00000338008.5
ENST00000510158.1 ENST00000505560.1 |
ZFYVE16
|
zinc finger, FYVE domain containing 16 |
| chr17_-_30185946 | 0.12 |
ENST00000579741.1
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr6_+_62284008 | 0.11 |
ENST00000544932.1
|
MTRNR2L9
|
MT-RNR2-like 9 (pseudogene) |
| chr3_-_64009658 | 0.11 |
ENST00000394431.2
|
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr17_+_67957878 | 0.11 |
ENST00000420427.1
|
AC004562.1
|
AC004562.1 |
| chr4_-_80994210 | 0.11 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr2_+_201171064 | 0.11 |
ENST00000451764.2
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr19_-_44160768 | 0.11 |
ENST00000593447.1
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr8_+_67344710 | 0.11 |
ENST00000379385.4
ENST00000396623.3 ENST00000415254.1 |
ADHFE1
|
alcohol dehydrogenase, iron containing, 1 |
| chr12_+_97306295 | 0.11 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr2_+_175260514 | 0.11 |
ENST00000424069.1
ENST00000427038.1 |
SCRN3
|
secernin 3 |
| chr12_-_75905374 | 0.11 |
ENST00000438169.2
ENST00000229214.4 |
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr6_-_80247105 | 0.11 |
ENST00000369846.4
|
LCA5
|
Leber congenital amaurosis 5 |
| chr12_+_56435637 | 0.11 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chr15_+_86087267 | 0.11 |
ENST00000558166.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr7_+_112063192 | 0.11 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr20_+_48552908 | 0.11 |
ENST00000244061.2
|
RNF114
|
ring finger protein 114 |
| chr14_+_32547434 | 0.11 |
ENST00000556191.1
ENST00000554090.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr12_-_76462713 | 0.11 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr16_-_24942411 | 0.11 |
ENST00000571843.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr2_+_201170770 | 0.11 |
ENST00000409988.3
ENST00000409385.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr4_-_111119804 | 0.11 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr10_+_124739911 | 0.10 |
ENST00000405485.1
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr4_-_14889791 | 0.10 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
| chr13_-_53024725 | 0.10 |
ENST00000378060.4
|
VPS36
|
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chr5_+_65222500 | 0.10 |
ENST00000511297.1
ENST00000506030.1 |
ERBB2IP
|
erbb2 interacting protein |
| chr1_-_168464875 | 0.10 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr7_+_142919130 | 0.10 |
ENST00000408947.3
|
TAS2R40
|
taste receptor, type 2, member 40 |
| chr2_-_201729284 | 0.10 |
ENST00000434813.2
|
CLK1
|
CDC-like kinase 1 |
| chr2_+_171640291 | 0.10 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr16_+_22517166 | 0.10 |
ENST00000356156.3
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr19_+_12721760 | 0.10 |
ENST00000600752.1
ENST00000540038.1 |
ZNF791
|
zinc finger protein 791 |
| chr21_+_34602377 | 0.10 |
ENST00000342101.3
ENST00000413881.1 ENST00000443073.1 |
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr1_+_174844645 | 0.10 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr5_+_32531893 | 0.10 |
ENST00000512913.1
|
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr8_-_93978357 | 0.10 |
ENST00000522925.1
ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr8_+_125860939 | 0.10 |
ENST00000525292.1
ENST00000528090.1 |
LINC00964
|
long intergenic non-protein coding RNA 964 |
| chr10_+_111967345 | 0.10 |
ENST00000332674.5
ENST00000453116.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr6_-_100016527 | 0.09 |
ENST00000523985.1
ENST00000518714.1 ENST00000520371.1 |
CCNC
|
cyclin C |
| chr9_+_127054254 | 0.09 |
ENST00000422297.1
|
NEK6
|
NIMA-related kinase 6 |
| chr12_+_32832134 | 0.09 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA2_FOXJ3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.1 | 0.5 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.1 | 0.3 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.4 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.2 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 0.3 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 0.2 | GO:1900244 | synaptic vesicle recycling via endosome(GO:0036466) positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.1 | 0.2 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.2 | GO:0086097 | renin-angiotensin regulation of aldosterone production(GO:0002018) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.3 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.0 | 0.3 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.2 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.3 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.2 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:0045423 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.1 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0006664 | glycolipid metabolic process(GO:0006664) liposaccharide metabolic process(GO:1903509) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.3 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:2000627 | rRNA import into mitochondrion(GO:0035928) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.3 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.3 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.9 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.0 | 0.1 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.2 | GO:0090309 | maintenance of DNA methylation(GO:0010216) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.3 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.2 | GO:0007379 | segment specification(GO:0007379) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.7 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.1 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.0 | GO:0072616 | interleukin-18 secretion(GO:0072616) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.3 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.0 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.0 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.0 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.3 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 1.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.2 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.0 | GO:0017076 | purine nucleotide binding(GO:0017076) |
| 0.0 | 0.4 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.1 | GO:0004877 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.0 | 0.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.4 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.2 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.0 | GO:0050473 | linoleate 13S-lipoxygenase activity(GO:0016165) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0060229 | lipase activator activity(GO:0060229) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.0 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |