Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
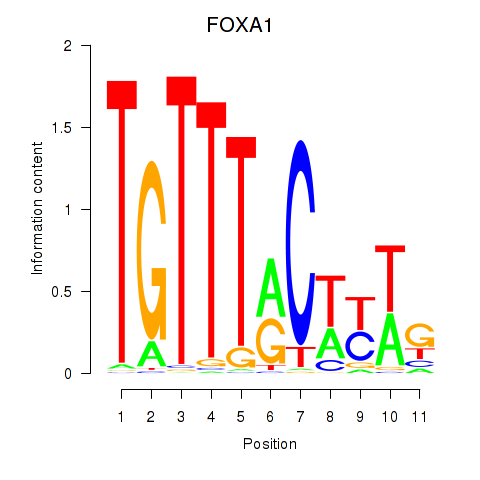
Results for FOXA1
Z-value: 0.37
Transcription factors associated with FOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA1
|
ENSG00000129514.4 | forkhead box A1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA1 | hg19_v2_chr14_-_38064198_38064239 | 0.41 | 5.9e-01 | Click! |
Activity profile of FOXA1 motif
Sorted Z-values of FOXA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_102200948 | 0.25 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr14_-_37051798 | 0.19 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr7_-_84121858 | 0.18 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr2_-_220042825 | 0.17 |
ENST00000409789.1
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr1_-_201096312 | 0.16 |
ENST00000449188.2
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chr6_+_26251835 | 0.15 |
ENST00000356350.2
|
HIST1H2BH
|
histone cluster 1, H2bh |
| chr15_+_101417919 | 0.15 |
ENST00000561338.1
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr19_+_18496957 | 0.14 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr19_-_45909585 | 0.14 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr17_+_70026795 | 0.14 |
ENST00000472655.2
ENST00000538810.1 |
RP11-84E24.3
|
long intergenic non-protein coding RNA 1152 |
| chr19_+_42806250 | 0.13 |
ENST00000598490.1
ENST00000341747.3 |
PRR19
|
proline rich 19 |
| chr17_-_26694979 | 0.13 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr11_-_47206965 | 0.13 |
ENST00000525725.1
|
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr2_-_220041860 | 0.13 |
ENST00000453038.1
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr17_-_26695013 | 0.12 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr17_+_79953310 | 0.12 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr19_-_30199516 | 0.12 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr10_+_104535994 | 0.11 |
ENST00000369889.4
|
WBP1L
|
WW domain binding protein 1-like |
| chr11_+_34999328 | 0.11 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr19_-_56047661 | 0.11 |
ENST00000344158.3
|
SBK2
|
SH3 domain binding kinase family, member 2 |
| chr2_-_175202151 | 0.11 |
ENST00000595354.1
|
AC018470.1
|
Uncharacterized protein FLJ46347 |
| chr17_+_72426891 | 0.11 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr21_+_17791648 | 0.11 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr11_+_844406 | 0.10 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr19_+_18492973 | 0.10 |
ENST00000595973.2
|
GDF15
|
growth differentiation factor 15 |
| chr5_+_149980622 | 0.10 |
ENST00000394243.1
|
SYNPO
|
synaptopodin |
| chr11_+_119205222 | 0.10 |
ENST00000311413.4
|
RNF26
|
ring finger protein 26 |
| chr19_+_44037334 | 0.09 |
ENST00000314228.5
|
ZNF575
|
zinc finger protein 575 |
| chr2_+_58655461 | 0.09 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr4_-_103940791 | 0.09 |
ENST00000510559.1
ENST00000394789.3 ENST00000296422.7 |
SLC9B1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr12_-_54778244 | 0.09 |
ENST00000549937.1
|
ZNF385A
|
zinc finger protein 385A |
| chr15_+_75335604 | 0.08 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr5_+_125706998 | 0.08 |
ENST00000506445.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr3_+_52245458 | 0.08 |
ENST00000459884.1
|
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr14_+_105267250 | 0.08 |
ENST00000342537.7
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr1_+_198126209 | 0.08 |
ENST00000367383.1
|
NEK7
|
NIMA-related kinase 7 |
| chr14_-_64971288 | 0.08 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr1_+_43855545 | 0.08 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr17_+_36873677 | 0.08 |
ENST00000471200.1
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr2_+_105050794 | 0.07 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr3_+_186692745 | 0.07 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr17_-_27038063 | 0.07 |
ENST00000439862.3
|
PROCA1
|
protein interacting with cyclin A1 |
| chr12_+_121416489 | 0.07 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr2_-_183291741 | 0.07 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr5_+_150157444 | 0.07 |
ENST00000526627.1
|
SMIM3
|
small integral membrane protein 3 |
| chr12_+_10365082 | 0.07 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr12_+_121416340 | 0.07 |
ENST00000257555.6
ENST00000400024.2 |
HNF1A
|
HNF1 homeobox A |
| chr1_+_227127981 | 0.07 |
ENST00000366778.1
ENST00000366777.3 ENST00000458507.2 |
ADCK3
|
aarF domain containing kinase 3 |
| chr16_+_4896659 | 0.07 |
ENST00000592120.1
|
UBN1
|
ubinuclein 1 |
| chr13_-_46716969 | 0.07 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr6_+_88054530 | 0.07 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr5_+_150157860 | 0.07 |
ENST00000600109.1
|
AC010441.1
|
AC010441.1 |
| chr12_-_71551868 | 0.07 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr7_+_134832808 | 0.07 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr16_-_4665023 | 0.07 |
ENST00000591897.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr10_-_90751038 | 0.06 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr1_+_61547894 | 0.06 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr20_+_306177 | 0.06 |
ENST00000544632.1
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr2_+_162101247 | 0.06 |
ENST00000439050.1
ENST00000436506.1 |
AC009299.3
|
AC009299.3 |
| chrX_+_37639302 | 0.06 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr7_-_84122033 | 0.06 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr4_+_165675269 | 0.06 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr21_+_33671264 | 0.06 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr4_+_75174180 | 0.06 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr7_+_106415457 | 0.06 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr18_-_2982869 | 0.06 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr17_+_72427477 | 0.06 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr19_-_48894104 | 0.06 |
ENST00000597017.1
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr2_-_233877912 | 0.05 |
ENST00000264051.3
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr1_-_177939041 | 0.05 |
ENST00000308284.6
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr12_-_56123444 | 0.05 |
ENST00000546457.1
ENST00000549117.1 |
CD63
|
CD63 molecule |
| chr17_-_19651668 | 0.05 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr17_-_46035187 | 0.05 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr12_-_54778471 | 0.05 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr3_+_137717571 | 0.05 |
ENST00000343735.4
|
CLDN18
|
claudin 18 |
| chr1_+_43855560 | 0.05 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr4_-_40516560 | 0.05 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr21_+_17792672 | 0.05 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr6_-_117747015 | 0.05 |
ENST00000368508.3
ENST00000368507.3 |
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase |
| chr17_-_17485731 | 0.05 |
ENST00000395783.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr18_+_60382672 | 0.05 |
ENST00000400316.4
ENST00000262719.5 |
PHLPP1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr20_+_306221 | 0.05 |
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr17_+_8924837 | 0.05 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr19_-_50380536 | 0.05 |
ENST00000391832.3
ENST00000391834.2 ENST00000344175.5 |
AKT1S1
|
AKT1 substrate 1 (proline-rich) |
| chr18_+_3449330 | 0.04 |
ENST00000549253.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr2_+_196522032 | 0.04 |
ENST00000418005.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr19_+_50380682 | 0.04 |
ENST00000221543.5
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr4_+_15376165 | 0.04 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr5_-_115872124 | 0.04 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr2_-_214016314 | 0.04 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr1_+_74701062 | 0.04 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr5_+_60933634 | 0.04 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr21_+_33671160 | 0.04 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr8_-_116681123 | 0.04 |
ENST00000519674.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chrX_+_37639264 | 0.04 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr11_+_35684288 | 0.04 |
ENST00000299413.5
|
TRIM44
|
tripartite motif containing 44 |
| chr7_+_28452130 | 0.04 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr11_+_1151573 | 0.04 |
ENST00000534821.1
ENST00000356191.2 |
MUC5AC
|
mucin 5AC, oligomeric mucus/gel-forming |
| chr17_+_74372662 | 0.04 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chr7_-_107968921 | 0.04 |
ENST00000442580.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr19_+_50380917 | 0.04 |
ENST00000535102.2
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr12_+_10365404 | 0.04 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr10_-_102890883 | 0.04 |
ENST00000445873.1
|
TLX1NB
|
TLX1 neighbor |
| chr10_-_75676400 | 0.03 |
ENST00000412307.2
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr2_-_87248975 | 0.03 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr8_-_30002179 | 0.03 |
ENST00000320542.3
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4 |
| chr15_+_28624878 | 0.03 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr3_+_186330712 | 0.03 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr10_-_21806759 | 0.03 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr20_-_43150601 | 0.03 |
ENST00000541235.1
ENST00000255175.1 ENST00000342374.4 |
SERINC3
|
serine incorporator 3 |
| chr14_+_23299088 | 0.03 |
ENST00000355151.5
ENST00000397496.3 ENST00000555345.1 ENST00000432849.3 ENST00000553711.1 ENST00000556465.1 ENST00000397505.2 ENST00000557221.1 ENST00000311892.6 ENST00000556840.1 ENST00000555536.1 |
MRPL52
|
mitochondrial ribosomal protein L52 |
| chr6_+_108977520 | 0.03 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr12_-_106697974 | 0.03 |
ENST00000553039.1
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr17_-_48546232 | 0.03 |
ENST00000258969.4
|
CHAD
|
chondroadherin |
| chr15_+_59910132 | 0.03 |
ENST00000559200.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr5_+_40841276 | 0.03 |
ENST00000254691.5
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr16_-_4664860 | 0.03 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr12_-_56122761 | 0.03 |
ENST00000552164.1
ENST00000420846.3 ENST00000257857.4 |
CD63
|
CD63 molecule |
| chr1_+_12538594 | 0.03 |
ENST00000543710.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr6_+_7108210 | 0.03 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr14_-_24911448 | 0.03 |
ENST00000555355.1
ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr12_+_96588368 | 0.03 |
ENST00000547860.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr16_-_4896205 | 0.03 |
ENST00000589389.1
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr6_+_7107830 | 0.03 |
ENST00000379933.3
|
RREB1
|
ras responsive element binding protein 1 |
| chr8_-_131028782 | 0.03 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr17_+_9728828 | 0.03 |
ENST00000262441.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr12_-_89746264 | 0.03 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chrX_+_24167746 | 0.03 |
ENST00000428571.1
ENST00000539115.1 |
ZFX
|
zinc finger protein, X-linked |
| chr16_+_73420942 | 0.03 |
ENST00000554640.1
ENST00000562661.1 ENST00000561875.1 |
RP11-140I24.1
|
RP11-140I24.1 |
| chr2_+_66918558 | 0.03 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr14_+_23654525 | 0.03 |
ENST00000399910.1
ENST00000492621.1 |
C14orf164
|
chromosome 14 open reading frame 164 |
| chr10_+_99400443 | 0.03 |
ENST00000370631.3
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr6_-_161085291 | 0.03 |
ENST00000316300.5
|
LPA
|
lipoprotein, Lp(a) |
| chr12_-_53343560 | 0.03 |
ENST00000548998.1
|
KRT8
|
keratin 8 |
| chr5_-_142065612 | 0.03 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr20_-_50179368 | 0.03 |
ENST00000609943.1
ENST00000609507.1 |
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
| chr8_-_116681221 | 0.02 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr16_+_21608525 | 0.02 |
ENST00000567404.1
|
METTL9
|
methyltransferase like 9 |
| chr19_+_39214797 | 0.02 |
ENST00000440400.1
|
ACTN4
|
actinin, alpha 4 |
| chr14_+_56585048 | 0.02 |
ENST00000267460.4
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr9_+_136287444 | 0.02 |
ENST00000355699.2
ENST00000356589.2 ENST00000371911.3 |
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr22_-_45404819 | 0.02 |
ENST00000447824.3
ENST00000404079.2 ENST00000420689.1 ENST00000403565.1 |
PHF21B
|
PHD finger protein 21B |
| chr18_-_53070913 | 0.02 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chrX_-_63425561 | 0.02 |
ENST00000374869.3
ENST00000330258.3 |
AMER1
|
APC membrane recruitment protein 1 |
| chr3_-_123123407 | 0.02 |
ENST00000466617.1
|
ADCY5
|
adenylate cyclase 5 |
| chrX_+_9431324 | 0.02 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chrX_-_124097620 | 0.02 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr12_+_103981044 | 0.02 |
ENST00000388887.2
|
STAB2
|
stabilin 2 |
| chr5_+_40841410 | 0.02 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr2_+_65283529 | 0.02 |
ENST00000546106.1
ENST00000537589.1 ENST00000260569.4 |
CEP68
|
centrosomal protein 68kDa |
| chr12_-_92536433 | 0.02 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chr6_-_107077347 | 0.02 |
ENST00000369063.3
ENST00000539449.1 |
RTN4IP1
|
reticulon 4 interacting protein 1 |
| chr6_+_7107999 | 0.02 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr3_+_160394940 | 0.02 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chr8_+_119294456 | 0.02 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr7_+_73242490 | 0.02 |
ENST00000431918.1
|
CLDN4
|
claudin 4 |
| chr7_+_95115210 | 0.02 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr9_+_105757590 | 0.02 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr11_-_9286921 | 0.02 |
ENST00000328194.3
|
DENND5A
|
DENN/MADD domain containing 5A |
| chr6_+_143929307 | 0.02 |
ENST00000427704.2
ENST00000305766.6 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr1_+_199996702 | 0.02 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr1_+_220701456 | 0.02 |
ENST00000366918.4
ENST00000402574.1 |
MARK1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr5_-_115872142 | 0.02 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr1_+_223101757 | 0.02 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr2_+_173724771 | 0.02 |
ENST00000538974.1
ENST00000540783.1 |
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_+_173792893 | 0.02 |
ENST00000535187.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr1_+_199996733 | 0.02 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr2_-_182545603 | 0.02 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr7_-_7782204 | 0.01 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr8_+_77593448 | 0.01 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr6_+_54172653 | 0.01 |
ENST00000370869.3
|
TINAG
|
tubulointerstitial nephritis antigen |
| chr3_-_129407535 | 0.01 |
ENST00000432054.2
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr6_-_43596899 | 0.01 |
ENST00000307126.5
ENST00000452781.1 |
GTPBP2
|
GTP binding protein 2 |
| chr6_+_47666275 | 0.01 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr5_-_41510656 | 0.01 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chrX_+_37865804 | 0.01 |
ENST00000297875.2
ENST00000357972.5 |
SYTL5
|
synaptotagmin-like 5 |
| chr1_-_115301235 | 0.01 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr10_-_49482907 | 0.01 |
ENST00000374201.3
ENST00000407470.4 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr3_+_137728842 | 0.01 |
ENST00000183605.5
|
CLDN18
|
claudin 18 |
| chr13_-_67802549 | 0.01 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr7_-_92855762 | 0.01 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr6_+_106546808 | 0.01 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr15_+_71389281 | 0.01 |
ENST00000355327.3
|
THSD4
|
thrombospondin, type I, domain containing 4 |
| chr2_+_71163051 | 0.01 |
ENST00000412314.1
|
ATP6V1B1
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1 |
| chr12_+_121416437 | 0.01 |
ENST00000402929.1
ENST00000535955.1 ENST00000538626.1 ENST00000543427.1 |
HNF1A
|
HNF1 homeobox A |
| chr12_+_40787194 | 0.01 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr10_+_24738355 | 0.01 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr3_+_147657764 | 0.01 |
ENST00000467198.1
ENST00000485006.1 |
RP11-71N10.1
|
RP11-71N10.1 |
| chr3_+_69985792 | 0.01 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr3_+_189349162 | 0.01 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chrX_-_20237059 | 0.01 |
ENST00000457145.1
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr8_-_74495065 | 0.01 |
ENST00000523533.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr1_-_1356719 | 0.01 |
ENST00000520296.1
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr1_-_246729544 | 0.01 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr21_+_17442799 | 0.01 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_+_87797351 | 0.01 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr3_+_148545586 | 0.01 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr4_+_86396321 | 0.01 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr6_+_33589161 | 0.01 |
ENST00000605930.1
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr17_-_40897043 | 0.01 |
ENST00000428826.2
ENST00000592492.1 ENST00000585893.1 ENST00000593214.1 ENST00000590078.1 ENST00000586382.1 ENST00000415827.2 ENST00000592743.1 ENST00000586089.1 ENST00000435174.1 |
EZH1
|
enhancer of zeste homolog 1 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.2 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.2 | GO:0001519 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.0 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.0 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0031783 | melanocortin receptor binding(GO:0031779) corticotropin hormone receptor binding(GO:0031780) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) type 5 melanocortin receptor binding(GO:0031783) type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.0 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |