Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
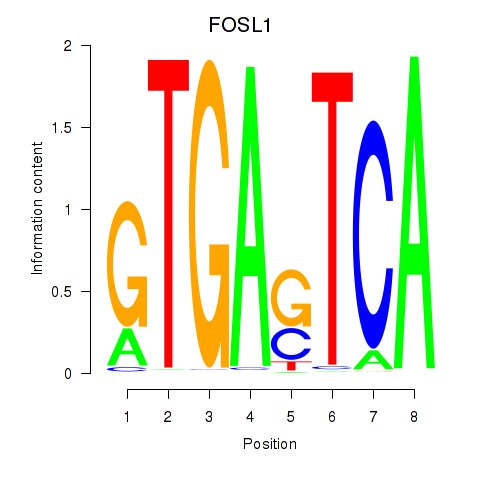
Results for FOSL1
Z-value: 0.53
Transcription factors associated with FOSL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOSL1
|
ENSG00000175592.4 | FOS like 1, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOSL1 | hg19_v2_chr11_-_65667884_65667895 | -0.82 | 1.8e-01 | Click! |
Activity profile of FOSL1 motif
Sorted Z-values of FOSL1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_58159361 | 0.39 |
ENST00000546567.1
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chr19_-_56056888 | 0.32 |
ENST00000592464.1
ENST00000420723.3 |
SBK3
|
SH3 domain binding kinase family, member 3 |
| chr11_-_119066545 | 0.25 |
ENST00000415318.1
|
CCDC153
|
coiled-coil domain containing 153 |
| chr6_+_138188351 | 0.24 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr7_-_130598059 | 0.23 |
ENST00000432045.2
|
MIR29B1
|
microRNA 29a |
| chr11_+_394145 | 0.23 |
ENST00000528036.1
|
PKP3
|
plakophilin 3 |
| chr17_+_74261413 | 0.22 |
ENST00000587913.1
|
UBALD2
|
UBA-like domain containing 2 |
| chr15_+_89182156 | 0.22 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr8_+_144821557 | 0.22 |
ENST00000534398.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr14_+_38065052 | 0.21 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr12_-_122658746 | 0.21 |
ENST00000377035.1
|
IL31
|
interleukin 31 |
| chr17_-_70417365 | 0.20 |
ENST00000580948.1
|
LINC00511
|
long intergenic non-protein coding RNA 511 |
| chr7_+_22766766 | 0.20 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr6_+_135818979 | 0.19 |
ENST00000421378.2
ENST00000579057.1 ENST00000436554.1 ENST00000438618.2 |
LINC00271
|
long intergenic non-protein coding RNA 271 |
| chr17_-_8021710 | 0.19 |
ENST00000380149.1
ENST00000448843.2 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr19_-_36019123 | 0.19 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr1_+_150480576 | 0.18 |
ENST00000346569.6
|
ECM1
|
extracellular matrix protein 1 |
| chr5_+_135394840 | 0.17 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr17_+_21191341 | 0.17 |
ENST00000526076.2
ENST00000361818.5 ENST00000316920.6 |
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr12_+_52695617 | 0.17 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr1_-_8000872 | 0.17 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr1_-_6659876 | 0.16 |
ENST00000496707.1
|
KLHL21
|
kelch-like family member 21 |
| chr11_+_393428 | 0.16 |
ENST00000533249.1
ENST00000527442.1 |
PKP3
|
plakophilin 3 |
| chr11_-_102668879 | 0.15 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr11_-_67141640 | 0.15 |
ENST00000533438.1
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr8_-_38386175 | 0.15 |
ENST00000437935.2
ENST00000358138.1 |
C8orf86
|
chromosome 8 open reading frame 86 |
| chr17_+_48712138 | 0.15 |
ENST00000515707.1
|
ABCC3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr19_-_40931891 | 0.15 |
ENST00000357949.4
|
SERTAD1
|
SERTA domain containing 1 |
| chr11_+_128563652 | 0.14 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_-_44233361 | 0.14 |
ENST00000275015.5
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr1_-_153935791 | 0.14 |
ENST00000429040.1
|
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr7_+_143078379 | 0.14 |
ENST00000449630.1
ENST00000457235.1 |
ZYX
|
zyxin |
| chr12_+_57914742 | 0.14 |
ENST00000551351.1
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr16_+_67312049 | 0.14 |
ENST00000565899.1
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr20_-_48782639 | 0.13 |
ENST00000435301.2
|
RP11-112L6.3
|
RP11-112L6.3 |
| chr9_+_97562440 | 0.13 |
ENST00000395357.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr11_-_62323702 | 0.13 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr19_+_46009837 | 0.13 |
ENST00000589627.1
|
VASP
|
vasodilator-stimulated phosphoprotein |
| chr12_+_57914481 | 0.13 |
ENST00000548887.1
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr12_-_57914275 | 0.13 |
ENST00000547303.1
ENST00000552740.1 ENST00000547526.1 ENST00000551116.1 ENST00000346473.3 |
DDIT3
|
DNA-damage-inducible transcript 3 |
| chr19_-_51523412 | 0.13 |
ENST00000391805.1
ENST00000599077.1 |
KLK10
|
kallikrein-related peptidase 10 |
| chr1_-_235116495 | 0.13 |
ENST00000549744.1
|
RP11-443B7.3
|
RP11-443B7.3 |
| chr17_+_74261277 | 0.13 |
ENST00000327490.6
|
UBALD2
|
UBA-like domain containing 2 |
| chr14_-_23426231 | 0.13 |
ENST00000556915.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr11_-_65150103 | 0.13 |
ENST00000294187.6
ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr11_-_66103932 | 0.12 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr1_-_207224307 | 0.12 |
ENST00000315927.4
|
YOD1
|
YOD1 deubiquitinase |
| chr15_+_89182178 | 0.12 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr12_+_48152774 | 0.12 |
ENST00000549243.1
|
SLC48A1
|
solute carrier family 48 (heme transporter), member 1 |
| chr6_+_151358048 | 0.12 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr15_+_91416092 | 0.12 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr9_+_140172200 | 0.12 |
ENST00000357503.2
|
TOR4A
|
torsin family 4, member A |
| chr2_+_87808725 | 0.12 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr9_-_35112376 | 0.12 |
ENST00000488109.2
|
FAM214B
|
family with sequence similarity 214, member B |
| chr11_+_46316677 | 0.12 |
ENST00000534787.1
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr1_-_95007193 | 0.12 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr16_+_30751953 | 0.12 |
ENST00000483578.1
|
RP11-2C24.4
|
RP11-2C24.4 |
| chr11_-_6341844 | 0.12 |
ENST00000303927.3
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr10_-_128110441 | 0.11 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr12_-_102591604 | 0.11 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr22_+_45072925 | 0.11 |
ENST00000006251.7
|
PRR5
|
proline rich 5 (renal) |
| chr14_+_103800513 | 0.11 |
ENST00000560338.1
ENST00000560763.1 ENST00000558316.1 ENST00000558265.1 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr11_+_35639735 | 0.11 |
ENST00000317811.4
|
FJX1
|
four jointed box 1 (Drosophila) |
| chr11_-_2924720 | 0.11 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr11_-_9025541 | 0.10 |
ENST00000525100.1
ENST00000309166.3 ENST00000531090.1 |
NRIP3
|
nuclear receptor interacting protein 3 |
| chr11_-_62457371 | 0.10 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr11_-_3862206 | 0.10 |
ENST00000351018.4
|
RHOG
|
ras homolog family member G |
| chr1_-_1009683 | 0.10 |
ENST00000453464.2
|
RNF223
|
ring finger protein 223 |
| chr11_+_57531292 | 0.10 |
ENST00000524579.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr4_-_185776789 | 0.10 |
ENST00000510284.1
|
RP11-701P16.5
|
RP11-701P16.5 |
| chr6_-_41673552 | 0.10 |
ENST00000419574.1
ENST00000445214.1 |
TFEB
|
transcription factor EB |
| chr9_+_140119618 | 0.10 |
ENST00000359069.2
|
C9orf169
|
chromosome 9 open reading frame 169 |
| chr7_+_150065278 | 0.10 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr17_+_76311791 | 0.10 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr1_-_153935738 | 0.10 |
ENST00000417348.1
|
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr2_+_87755054 | 0.10 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr11_+_394196 | 0.10 |
ENST00000331563.2
ENST00000531857.1 |
PKP3
|
plakophilin 3 |
| chr1_+_35220613 | 0.10 |
ENST00000338513.1
|
GJB5
|
gap junction protein, beta 5, 31.1kDa |
| chr1_+_33722080 | 0.09 |
ENST00000483388.1
ENST00000539719.1 |
ZNF362
|
zinc finger protein 362 |
| chr5_-_141061759 | 0.09 |
ENST00000508305.1
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr17_-_27044903 | 0.09 |
ENST00000395245.3
|
RAB34
|
RAB34, member RAS oncogene family |
| chr6_-_36410620 | 0.09 |
ENST00000454782.2
|
PXT1
|
peroxisomal, testis specific 1 |
| chr3_-_196065374 | 0.09 |
ENST00000454715.1
|
TM4SF19
|
transmembrane 4 L six family member 19 |
| chrX_-_153718953 | 0.09 |
ENST00000369649.4
ENST00000393586.1 |
SLC10A3
|
solute carrier family 10, member 3 |
| chr11_-_66103867 | 0.09 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr16_+_30077055 | 0.09 |
ENST00000564595.2
ENST00000569798.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr12_+_13349711 | 0.09 |
ENST00000538364.1
ENST00000396301.3 |
EMP1
|
epithelial membrane protein 1 |
| chr12_+_6644443 | 0.09 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_+_150480551 | 0.09 |
ENST00000369049.4
ENST00000369047.4 |
ECM1
|
extracellular matrix protein 1 |
| chr1_+_156084461 | 0.09 |
ENST00000347559.2
ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA
|
lamin A/C |
| chr17_-_27405875 | 0.09 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr12_+_9142131 | 0.09 |
ENST00000356986.3
ENST00000266551.4 |
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr9_+_131902346 | 0.09 |
ENST00000432124.1
ENST00000435305.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr11_-_66104237 | 0.09 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr14_-_68157084 | 0.09 |
ENST00000557564.1
|
RP11-1012A1.4
|
RP11-1012A1.4 |
| chr3_+_48507210 | 0.09 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr11_-_57417405 | 0.08 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr1_-_117753540 | 0.08 |
ENST00000328189.3
ENST00000369458.3 |
VTCN1
|
V-set domain containing T cell activation inhibitor 1 |
| chr19_-_46285646 | 0.08 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr16_+_30418910 | 0.08 |
ENST00000566625.1
|
ZNF771
|
zinc finger protein 771 |
| chr15_-_44828838 | 0.08 |
ENST00000560750.1
|
EIF3J-AS1
|
EIF3J antisense RNA 1 (head to head) |
| chr20_+_48429356 | 0.08 |
ENST00000361573.2
ENST00000541138.1 ENST00000539601.1 |
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr17_-_45928521 | 0.08 |
ENST00000536300.1
|
SP6
|
Sp6 transcription factor |
| chr11_-_6341724 | 0.08 |
ENST00000530979.1
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr12_+_56546363 | 0.08 |
ENST00000551834.1
ENST00000552568.1 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr14_-_23426322 | 0.08 |
ENST00000555367.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr17_+_28256874 | 0.08 |
ENST00000541045.1
ENST00000536908.2 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr17_+_80317121 | 0.08 |
ENST00000333437.4
|
TEX19
|
testis expressed 19 |
| chr7_+_73245193 | 0.08 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr11_+_46402297 | 0.08 |
ENST00000405308.2
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr22_+_42372764 | 0.08 |
ENST00000396426.3
ENST00000406029.1 |
SEPT3
|
septin 3 |
| chr12_+_13349650 | 0.08 |
ENST00000256951.5
ENST00000431267.2 ENST00000542474.1 ENST00000544053.1 |
EMP1
|
epithelial membrane protein 1 |
| chr9_-_91793675 | 0.08 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr17_-_39781054 | 0.08 |
ENST00000463128.1
|
KRT17
|
keratin 17 |
| chr22_+_45072958 | 0.08 |
ENST00000403581.1
|
PRR5
|
proline rich 5 (renal) |
| chrX_-_154563889 | 0.08 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr1_+_12042015 | 0.07 |
ENST00000412236.1
|
MFN2
|
mitofusin 2 |
| chr19_+_50432400 | 0.07 |
ENST00000423777.2
ENST00000600336.1 ENST00000597227.1 |
ATF5
|
activating transcription factor 5 |
| chr17_-_27044810 | 0.07 |
ENST00000395242.2
|
RAB34
|
RAB34, member RAS oncogene family |
| chr12_-_53625958 | 0.07 |
ENST00000327550.3
ENST00000546717.1 ENST00000425354.2 ENST00000394426.1 |
RARG
|
retinoic acid receptor, gamma |
| chr11_-_65149422 | 0.07 |
ENST00000526432.1
ENST00000527174.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr11_-_2950642 | 0.07 |
ENST00000314222.4
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2 |
| chr19_+_2476116 | 0.07 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr17_-_27045165 | 0.07 |
ENST00000436730.3
ENST00000450529.1 ENST00000583538.1 ENST00000419712.3 ENST00000580843.2 ENST00000582934.1 ENST00000415040.2 ENST00000353676.5 ENST00000453384.3 ENST00000447716.1 |
RAB34
|
RAB34, member RAS oncogene family |
| chr9_-_139948468 | 0.07 |
ENST00000312665.5
|
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2 |
| chr17_-_27045427 | 0.07 |
ENST00000301043.6
ENST00000412625.1 |
RAB34
|
RAB34, member RAS oncogene family |
| chr16_+_57662419 | 0.07 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr17_-_79481666 | 0.07 |
ENST00000575659.1
|
ACTG1
|
actin, gamma 1 |
| chr3_-_32022733 | 0.07 |
ENST00000438237.2
ENST00000396556.2 |
OSBPL10
|
oxysterol binding protein-like 10 |
| chr11_+_64126671 | 0.07 |
ENST00000530504.1
|
RPS6KA4
|
ribosomal protein S6 kinase, 90kDa, polypeptide 4 |
| chr1_+_27719148 | 0.07 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chr1_+_155107820 | 0.07 |
ENST00000484157.1
|
SLC50A1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr19_+_3366547 | 0.07 |
ENST00000341919.3
ENST00000590282.1 ENST00000443272.2 |
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor) |
| chr8_+_22844995 | 0.07 |
ENST00000524077.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr3_+_183894566 | 0.07 |
ENST00000439647.1
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chrX_-_153285251 | 0.07 |
ENST00000444230.1
ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr17_-_18908040 | 0.07 |
ENST00000388995.6
|
FAM83G
|
family with sequence similarity 83, member G |
| chr5_-_140013224 | 0.07 |
ENST00000498971.2
|
CD14
|
CD14 molecule |
| chr16_+_88519669 | 0.06 |
ENST00000319555.3
|
ZFPM1
|
zinc finger protein, FOG family member 1 |
| chr15_+_40531243 | 0.06 |
ENST00000558055.1
ENST00000455577.2 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr15_+_89181974 | 0.06 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr21_-_43786634 | 0.06 |
ENST00000291527.2
|
TFF1
|
trefoil factor 1 |
| chr19_+_50431959 | 0.06 |
ENST00000595125.1
|
ATF5
|
activating transcription factor 5 |
| chr2_-_220117867 | 0.06 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr1_-_160832642 | 0.06 |
ENST00000368034.4
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4 |
| chr1_+_17634689 | 0.06 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr5_+_55354822 | 0.06 |
ENST00000511861.1
|
CTD-2227I18.1
|
CTD-2227I18.1 |
| chr3_+_112930373 | 0.06 |
ENST00000498710.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr12_-_120805872 | 0.06 |
ENST00000546985.1
|
MSI1
|
musashi RNA-binding protein 1 |
| chr3_-_18466787 | 0.06 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr16_+_89988259 | 0.06 |
ENST00000554444.1
ENST00000556565.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr10_+_75668916 | 0.06 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr15_+_75335604 | 0.06 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr5_-_141061777 | 0.06 |
ENST00000239440.4
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr20_+_48429233 | 0.06 |
ENST00000417961.1
|
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr17_+_4853442 | 0.06 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr3_+_19988736 | 0.06 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr9_-_35111570 | 0.06 |
ENST00000378561.1
ENST00000603301.1 |
FAM214B
|
family with sequence similarity 214, member B |
| chr19_+_36630855 | 0.06 |
ENST00000589146.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr14_+_73525265 | 0.06 |
ENST00000525161.1
|
RBM25
|
RNA binding motif protein 25 |
| chr1_+_183155373 | 0.06 |
ENST00000493293.1
ENST00000264144.4 |
LAMC2
|
laminin, gamma 2 |
| chr16_+_28875126 | 0.06 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr1_-_155006224 | 0.06 |
ENST00000368424.3
|
DCST2
|
DC-STAMP domain containing 2 |
| chr3_+_69812877 | 0.06 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr1_-_153521714 | 0.06 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr7_+_28725585 | 0.06 |
ENST00000396298.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr1_-_204121102 | 0.06 |
ENST00000367202.4
|
ETNK2
|
ethanolamine kinase 2 |
| chr17_+_28268623 | 0.06 |
ENST00000394835.3
ENST00000320856.5 ENST00000394832.2 ENST00000378738.3 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr12_-_6809958 | 0.06 |
ENST00000320591.5
ENST00000534837.1 |
PIANP
|
PILR alpha associated neural protein |
| chr11_-_2924970 | 0.06 |
ENST00000533594.1
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr3_-_87040259 | 0.06 |
ENST00000383698.3
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr12_-_54813229 | 0.05 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr4_+_74606223 | 0.05 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr17_-_73150629 | 0.05 |
ENST00000356033.4
ENST00000405458.3 ENST00000409753.3 |
HN1
|
hematological and neurological expressed 1 |
| chr11_+_706219 | 0.05 |
ENST00000533500.1
|
EPS8L2
|
EPS8-like 2 |
| chr15_-_83315874 | 0.05 |
ENST00000569257.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chrX_-_129244454 | 0.05 |
ENST00000308167.5
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chr16_-_122619 | 0.05 |
ENST00000262316.6
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila) |
| chr3_+_69928256 | 0.05 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chr11_+_6411636 | 0.05 |
ENST00000299397.3
ENST00000356761.2 ENST00000342245.4 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr12_-_108991812 | 0.05 |
ENST00000392806.3
ENST00000547567.1 |
TMEM119
|
transmembrane protein 119 |
| chr7_+_48128194 | 0.05 |
ENST00000416681.1
ENST00000331803.4 ENST00000432131.1 |
UPP1
|
uridine phosphorylase 1 |
| chr10_+_17270214 | 0.05 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr14_-_64970494 | 0.05 |
ENST00000608382.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr17_+_65375082 | 0.05 |
ENST00000584471.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr17_-_27045405 | 0.05 |
ENST00000430132.2
|
RAB34
|
RAB34, member RAS oncogene family |
| chr15_-_83316711 | 0.05 |
ENST00000568128.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr12_+_120972606 | 0.05 |
ENST00000413266.2
|
RNF10
|
ring finger protein 10 |
| chr16_+_57662138 | 0.05 |
ENST00000562414.1
ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr15_-_44829057 | 0.05 |
ENST00000559356.1
ENST00000560049.1 ENST00000313807.4 |
EIF3J-AS1
|
EIF3J antisense RNA 1 (head to head) |
| chr16_+_28875268 | 0.05 |
ENST00000395532.4
|
SH2B1
|
SH2B adaptor protein 1 |
| chr1_+_151584544 | 0.05 |
ENST00000458013.2
ENST00000368843.3 |
SNX27
|
sorting nexin family member 27 |
| chr8_-_108510224 | 0.05 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr1_-_209824643 | 0.05 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr18_+_61143994 | 0.05 |
ENST00000382771.4
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr1_-_67142710 | 0.05 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr14_+_21467414 | 0.05 |
ENST00000554422.1
ENST00000298681.4 |
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr17_-_3461092 | 0.05 |
ENST00000301365.4
ENST00000572519.1 |
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3 |
| chr9_+_34992846 | 0.05 |
ENST00000443266.1
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chrX_-_153285395 | 0.05 |
ENST00000369980.3
|
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOSL1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.2 | GO:0072573 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 0.5 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.2 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.2 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.2 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.4 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.1 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.0 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.2 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0052042 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.0 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.0 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.2 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.0 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.0 | 0.1 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.0 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.1 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.2 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.0 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.0 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |