Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
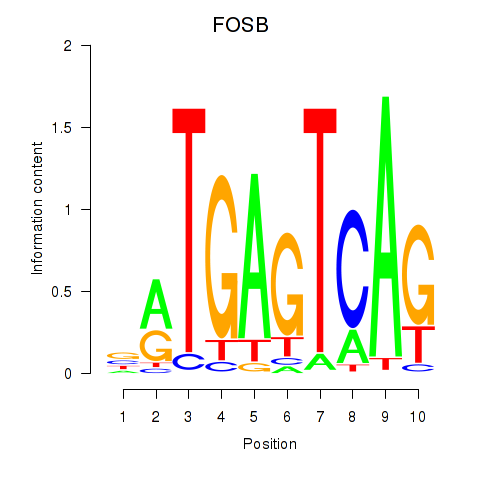
Results for FOSB
Z-value: 0.66
Transcription factors associated with FOSB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOSB
|
ENSG00000125740.9 | FosB proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOSB | hg19_v2_chr19_+_45973360_45973523 | -0.95 | 5.1e-02 | Click! |
Activity profile of FOSB motif
Sorted Z-values of FOSB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_56056888 | 0.70 |
ENST00000592464.1
ENST00000420723.3 |
SBK3
|
SH3 domain binding kinase family, member 3 |
| chr5_-_176923803 | 0.63 |
ENST00000506161.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr11_+_394145 | 0.60 |
ENST00000528036.1
|
PKP3
|
plakophilin 3 |
| chr19_-_46145696 | 0.45 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr17_+_7123207 | 0.41 |
ENST00000584103.1
ENST00000579886.2 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr1_-_153935791 | 0.40 |
ENST00000429040.1
|
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr5_-_176923846 | 0.39 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr16_+_30675654 | 0.33 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr3_-_48632593 | 0.33 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr14_-_75083313 | 0.33 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr8_+_125283924 | 0.31 |
ENST00000517482.1
|
RP11-383J24.1
|
RP11-383J24.1 |
| chr3_+_14474178 | 0.30 |
ENST00000452775.1
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr5_+_175288631 | 0.29 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr17_+_72426891 | 0.28 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr1_-_153935738 | 0.28 |
ENST00000417348.1
|
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr22_-_39151947 | 0.27 |
ENST00000216064.4
|
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr5_-_175843524 | 0.27 |
ENST00000502877.1
|
CLTB
|
clathrin, light chain B |
| chr3_-_48601206 | 0.26 |
ENST00000273610.3
|
UCN2
|
urocortin 2 |
| chr5_-_139930713 | 0.24 |
ENST00000602657.1
|
SRA1
|
steroid receptor RNA activator 1 |
| chr1_-_153521714 | 0.24 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr17_-_25568687 | 0.23 |
ENST00000581944.1
|
RP11-663N22.1
|
RP11-663N22.1 |
| chr9_+_131902283 | 0.23 |
ENST00000436883.1
ENST00000414510.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr11_+_394196 | 0.23 |
ENST00000331563.2
ENST00000531857.1 |
PKP3
|
plakophilin 3 |
| chr12_-_53625958 | 0.23 |
ENST00000327550.3
ENST00000546717.1 ENST00000425354.2 ENST00000394426.1 |
RARG
|
retinoic acid receptor, gamma |
| chr1_-_6662919 | 0.22 |
ENST00000377658.4
ENST00000377663.3 |
KLHL21
|
kelch-like family member 21 |
| chr16_+_30751953 | 0.22 |
ENST00000483578.1
|
RP11-2C24.4
|
RP11-2C24.4 |
| chr16_+_28875268 | 0.21 |
ENST00000395532.4
|
SH2B1
|
SH2B adaptor protein 1 |
| chr22_+_37959647 | 0.21 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr7_+_143078379 | 0.21 |
ENST00000449630.1
ENST00000457235.1 |
ZYX
|
zyxin |
| chr17_+_75315654 | 0.21 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr1_-_21059029 | 0.20 |
ENST00000444387.2
ENST00000375031.1 ENST00000518294.1 |
SH2D5
|
SH2 domain containing 5 |
| chr17_-_18908040 | 0.20 |
ENST00000388995.6
|
FAM83G
|
family with sequence similarity 83, member G |
| chr8_+_99076750 | 0.20 |
ENST00000545282.1
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr17_+_25799008 | 0.20 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr2_+_28615669 | 0.19 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr5_+_139493665 | 0.19 |
ENST00000331327.3
|
PURA
|
purine-rich element binding protein A |
| chr11_-_65150103 | 0.19 |
ENST00000294187.6
ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr18_-_74207146 | 0.19 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr16_+_28875126 | 0.18 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr14_-_23426322 | 0.18 |
ENST00000555367.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr16_-_1429674 | 0.18 |
ENST00000403703.1
ENST00000397464.1 ENST00000402641.2 |
UNKL
|
unkempt family zinc finger-like |
| chr3_-_149293990 | 0.18 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr22_-_39151434 | 0.17 |
ENST00000439339.1
|
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr1_-_153521597 | 0.17 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr6_-_30685214 | 0.17 |
ENST00000425072.1
|
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr11_-_66103932 | 0.17 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr15_+_67458357 | 0.17 |
ENST00000537194.2
|
SMAD3
|
SMAD family member 3 |
| chr1_-_235116495 | 0.17 |
ENST00000549744.1
|
RP11-443B7.3
|
RP11-443B7.3 |
| chr11_-_47206965 | 0.17 |
ENST00000525725.1
|
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr22_-_32058416 | 0.17 |
ENST00000439502.2
|
PISD
|
phosphatidylserine decarboxylase |
| chr1_-_21948906 | 0.17 |
ENST00000374761.2
ENST00000599760.1 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr22_-_32058166 | 0.17 |
ENST00000435900.1
ENST00000336566.4 |
PISD
|
phosphatidylserine decarboxylase |
| chr16_-_1429627 | 0.16 |
ENST00000248104.7
|
UNKL
|
unkempt family zinc finger-like |
| chr7_+_150065278 | 0.16 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr7_+_100728720 | 0.16 |
ENST00000306085.6
ENST00000412507.1 |
TRIM56
|
tripartite motif containing 56 |
| chr7_+_2687173 | 0.16 |
ENST00000403167.1
|
TTYH3
|
tweety family member 3 |
| chr12_-_13105081 | 0.16 |
ENST00000541128.1
|
GPRC5D
|
G protein-coupled receptor, family C, group 5, member D |
| chr9_+_131902346 | 0.16 |
ENST00000432124.1
ENST00000435305.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr1_+_154975110 | 0.15 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr16_-_4665023 | 0.15 |
ENST00000591897.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr12_+_53693466 | 0.15 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr17_+_46970134 | 0.15 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr17_-_79881408 | 0.15 |
ENST00000392366.3
|
MAFG
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr3_+_9958758 | 0.14 |
ENST00000383812.4
ENST00000438091.1 ENST00000295981.3 ENST00000436503.1 ENST00000403601.3 ENST00000416074.2 ENST00000455057.1 |
IL17RC
|
interleukin 17 receptor C |
| chr17_+_74733744 | 0.14 |
ENST00000586689.1
ENST00000587661.1 ENST00000593181.1 ENST00000336509.4 ENST00000355954.3 |
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr16_+_57662596 | 0.14 |
ENST00000567397.1
ENST00000568979.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr19_+_1248547 | 0.14 |
ENST00000586757.1
ENST00000300952.2 |
MIDN
|
midnolin |
| chr17_-_72864739 | 0.14 |
ENST00000579893.1
ENST00000544854.1 |
FDXR
|
ferredoxin reductase |
| chr1_-_24126051 | 0.14 |
ENST00000445705.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr14_-_23426231 | 0.14 |
ENST00000556915.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr11_-_66103867 | 0.13 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr15_+_101459420 | 0.13 |
ENST00000388948.3
ENST00000284395.5 ENST00000534045.1 ENST00000532029.2 |
LRRK1
|
leucine-rich repeat kinase 1 |
| chr17_-_27405875 | 0.13 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr17_-_27044903 | 0.13 |
ENST00000395245.3
|
RAB34
|
RAB34, member RAS oncogene family |
| chr17_-_27044810 | 0.13 |
ENST00000395242.2
|
RAB34
|
RAB34, member RAS oncogene family |
| chr3_+_9958870 | 0.13 |
ENST00000413608.1
|
IL17RC
|
interleukin 17 receptor C |
| chr18_+_3450161 | 0.13 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr9_-_21335356 | 0.13 |
ENST00000359039.4
|
KLHL9
|
kelch-like family member 9 |
| chr8_+_87111059 | 0.13 |
ENST00000285393.3
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr11_-_119066545 | 0.12 |
ENST00000415318.1
|
CCDC153
|
coiled-coil domain containing 153 |
| chr10_+_1102303 | 0.12 |
ENST00000381329.1
|
WDR37
|
WD repeat domain 37 |
| chr3_-_53878644 | 0.12 |
ENST00000481668.1
ENST00000467802.1 |
CHDH
|
choline dehydrogenase |
| chr17_+_9745786 | 0.12 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr15_+_75335604 | 0.12 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr11_+_128563652 | 0.12 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr7_+_871559 | 0.12 |
ENST00000421580.1
|
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr5_+_135394840 | 0.12 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr4_+_22999152 | 0.12 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr19_+_926000 | 0.12 |
ENST00000263620.3
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr17_+_80317121 | 0.12 |
ENST00000333437.4
|
TEX19
|
testis expressed 19 |
| chr15_+_41062159 | 0.11 |
ENST00000344320.6
|
C15orf62
|
chromosome 15 open reading frame 62 |
| chr12_-_49351303 | 0.11 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chr17_+_46970127 | 0.11 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr8_+_123875624 | 0.11 |
ENST00000534247.1
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr19_-_44008863 | 0.11 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3 |
| chr3_-_51994694 | 0.11 |
ENST00000395014.2
|
PCBP4
|
poly(rC) binding protein 4 |
| chr22_-_33968239 | 0.11 |
ENST00000452586.2
ENST00000421768.1 |
LARGE
|
like-glycosyltransferase |
| chr11_-_75201791 | 0.11 |
ENST00000529721.1
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr1_+_223889310 | 0.11 |
ENST00000434648.1
|
CAPN2
|
calpain 2, (m/II) large subunit |
| chr20_-_634000 | 0.11 |
ENST00000381962.3
|
SRXN1
|
sulfiredoxin 1 |
| chr15_+_45938079 | 0.10 |
ENST00000561493.1
|
SQRDL
|
sulfide quinone reductase-like (yeast) |
| chr5_-_66942617 | 0.10 |
ENST00000507298.1
|
RP11-83M16.5
|
RP11-83M16.5 |
| chr1_-_24126023 | 0.10 |
ENST00000429356.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr15_+_67418047 | 0.10 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr12_+_7023491 | 0.10 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chrX_-_153285251 | 0.10 |
ENST00000444230.1
ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr14_-_75536182 | 0.10 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr11_+_68671310 | 0.10 |
ENST00000255078.3
ENST00000539224.1 |
IGHMBP2
|
immunoglobulin mu binding protein 2 |
| chr8_-_17752996 | 0.10 |
ENST00000381841.2
ENST00000427924.1 |
FGL1
|
fibrinogen-like 1 |
| chr6_+_151358048 | 0.09 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr2_-_97509749 | 0.09 |
ENST00000331001.2
ENST00000318357.4 |
ANKRD23
|
ankyrin repeat domain 23 |
| chr12_+_57849048 | 0.09 |
ENST00000266646.2
|
INHBE
|
inhibin, beta E |
| chr16_-_57798253 | 0.09 |
ENST00000565270.1
|
KIFC3
|
kinesin family member C3 |
| chr9_-_73029540 | 0.09 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr1_+_154975258 | 0.09 |
ENST00000417934.2
|
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr15_+_69373210 | 0.09 |
ENST00000435479.1
ENST00000559870.1 |
LINC00277
RP11-809H16.5
|
long intergenic non-protein coding RNA 277 RP11-809H16.5 |
| chr12_+_58176525 | 0.09 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr11_+_28724129 | 0.09 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr10_+_17270214 | 0.08 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr2_+_69201705 | 0.08 |
ENST00000377938.2
|
GKN1
|
gastrokine 1 |
| chr22_+_23487513 | 0.08 |
ENST00000263116.2
ENST00000341989.4 |
RAB36
|
RAB36, member RAS oncogene family |
| chr17_-_27045165 | 0.08 |
ENST00000436730.3
ENST00000450529.1 ENST00000583538.1 ENST00000419712.3 ENST00000580843.2 ENST00000582934.1 ENST00000415040.2 ENST00000353676.5 ENST00000453384.3 ENST00000447716.1 |
RAB34
|
RAB34, member RAS oncogene family |
| chr19_-_46627914 | 0.08 |
ENST00000341415.2
|
IGFL3
|
IGF-like family member 3 |
| chr17_-_27467418 | 0.08 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr5_-_138534071 | 0.08 |
ENST00000394817.2
|
SIL1
|
SIL1 nucleotide exchange factor |
| chr15_+_59903975 | 0.08 |
ENST00000560585.1
ENST00000396065.1 |
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr19_-_55881741 | 0.08 |
ENST00000264563.2
ENST00000590625.1 ENST00000585513.1 |
IL11
|
interleukin 11 |
| chr12_+_28605426 | 0.08 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr12_+_6644443 | 0.08 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr11_-_9781068 | 0.08 |
ENST00000500698.1
|
RP11-540A21.2
|
RP11-540A21.2 |
| chr1_-_108231101 | 0.08 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr5_+_55354822 | 0.08 |
ENST00000511861.1
|
CTD-2227I18.1
|
CTD-2227I18.1 |
| chr14_+_103800513 | 0.08 |
ENST00000560338.1
ENST00000560763.1 ENST00000558316.1 ENST00000558265.1 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr12_+_10366016 | 0.08 |
ENST00000546017.1
ENST00000535576.1 ENST00000539170.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr19_-_18632861 | 0.08 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr16_+_57662419 | 0.08 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr17_+_79650962 | 0.08 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr9_+_137218362 | 0.08 |
ENST00000481739.1
|
RXRA
|
retinoid X receptor, alpha |
| chr16_+_57662138 | 0.08 |
ENST00000562414.1
ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr2_-_85641162 | 0.07 |
ENST00000447219.2
ENST00000409670.1 ENST00000409724.1 |
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr17_-_27045427 | 0.07 |
ENST00000301043.6
ENST00000412625.1 |
RAB34
|
RAB34, member RAS oncogene family |
| chr10_+_18549645 | 0.07 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr11_-_71781096 | 0.07 |
ENST00000535087.1
ENST00000535838.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr3_+_48507210 | 0.07 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr19_-_19739321 | 0.07 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr12_+_7023735 | 0.07 |
ENST00000538763.1
ENST00000544774.1 ENST00000545045.2 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr12_-_51402984 | 0.07 |
ENST00000545993.2
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr6_+_43027595 | 0.07 |
ENST00000259708.3
ENST00000472792.1 ENST00000479388.1 ENST00000460283.1 ENST00000394056.2 |
KLC4
|
kinesin light chain 4 |
| chr7_+_99647411 | 0.07 |
ENST00000438937.1
|
ZSCAN21
|
zinc finger and SCAN domain containing 21 |
| chr11_-_8739566 | 0.07 |
ENST00000533020.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr19_+_47104493 | 0.07 |
ENST00000291295.9
ENST00000597743.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chr16_-_4664860 | 0.07 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr17_+_74734052 | 0.07 |
ENST00000590514.1
|
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr4_+_48485341 | 0.07 |
ENST00000273861.4
|
SLC10A4
|
solute carrier family 10, member 4 |
| chr16_-_67281413 | 0.07 |
ENST00000258201.4
|
FHOD1
|
formin homology 2 domain containing 1 |
| chr2_-_11606275 | 0.07 |
ENST00000381525.3
ENST00000362009.4 |
E2F6
|
E2F transcription factor 6 |
| chr17_-_27503770 | 0.07 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr15_+_69373184 | 0.07 |
ENST00000558147.1
ENST00000440444.1 |
LINC00277
|
long intergenic non-protein coding RNA 277 |
| chr17_-_79481666 | 0.07 |
ENST00000575659.1
|
ACTG1
|
actin, gamma 1 |
| chr7_+_16700806 | 0.07 |
ENST00000446596.1
ENST00000438834.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr2_+_135596180 | 0.07 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr9_+_44867571 | 0.07 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr17_+_75315534 | 0.06 |
ENST00000590294.1
ENST00000329047.8 |
SEPT9
|
septin 9 |
| chr1_+_165796753 | 0.06 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr14_-_23426337 | 0.06 |
ENST00000342454.8
ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr8_+_144821557 | 0.06 |
ENST00000534398.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr2_+_220436917 | 0.06 |
ENST00000243786.2
|
INHA
|
inhibin, alpha |
| chr17_-_39942940 | 0.06 |
ENST00000310706.5
ENST00000393931.3 ENST00000424457.1 ENST00000591690.1 |
JUP
|
junction plakoglobin |
| chr1_+_27668505 | 0.06 |
ENST00000318074.5
|
SYTL1
|
synaptotagmin-like 1 |
| chr17_-_38721711 | 0.06 |
ENST00000578085.1
ENST00000246657.2 |
CCR7
|
chemokine (C-C motif) receptor 7 |
| chr9_+_140135665 | 0.06 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chr7_-_130598059 | 0.06 |
ENST00000432045.2
|
MIR29B1
|
microRNA 29a |
| chr20_+_48429233 | 0.06 |
ENST00000417961.1
|
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr6_+_32121218 | 0.06 |
ENST00000414204.1
ENST00000361568.2 ENST00000395523.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr17_+_46970178 | 0.06 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr14_-_91710852 | 0.06 |
ENST00000535815.1
ENST00000529102.1 |
GPR68
|
G protein-coupled receptor 68 |
| chr14_+_32476072 | 0.06 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chr15_-_76304731 | 0.05 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr12_-_71182695 | 0.05 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr4_+_108746282 | 0.05 |
ENST00000503862.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr14_+_97925151 | 0.05 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr15_+_75498739 | 0.05 |
ENST00000565074.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr5_-_41794663 | 0.05 |
ENST00000510634.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr4_-_89079817 | 0.05 |
ENST00000505480.1
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr1_+_56880606 | 0.05 |
ENST00000451914.1
|
RP4-710M16.2
|
RP4-710M16.2 |
| chr17_+_4853442 | 0.05 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr16_+_2083265 | 0.05 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr17_-_59668550 | 0.05 |
ENST00000521764.1
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2 |
| chr16_-_30125177 | 0.05 |
ENST00000406256.3
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr12_-_53297432 | 0.05 |
ENST00000546900.1
|
KRT8
|
keratin 8 |
| chr3_+_48507621 | 0.05 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chr5_+_43602750 | 0.05 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr11_-_2924720 | 0.05 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr19_+_34850385 | 0.05 |
ENST00000587521.2
ENST00000587384.1 ENST00000592277.1 |
GPI
|
glucose-6-phosphate isomerase |
| chr2_-_220083076 | 0.05 |
ENST00000295750.4
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr15_+_89631381 | 0.05 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr7_+_1084206 | 0.05 |
ENST00000444847.1
|
GPR146
|
G protein-coupled receptor 146 |
| chr9_-_21335240 | 0.05 |
ENST00000537938.1
|
KLHL9
|
kelch-like family member 9 |
| chr14_-_54317532 | 0.05 |
ENST00000418927.1
|
AL162759.1
|
AL162759.1 |
| chr19_-_35992780 | 0.05 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOSB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.2 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.1 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.4 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.3 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.1 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.5 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:0030473 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) |
| 0.0 | 0.2 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.4 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.4 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.4 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0052330 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.0 | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.1 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.3 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.6 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.2 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.4 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.3 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0004324 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.0 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.0 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.0 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |