Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
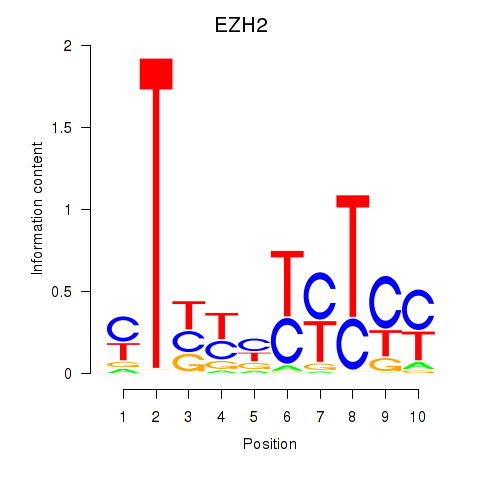
Results for EZH2
Z-value: 1.43
Transcription factors associated with EZH2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EZH2
|
ENSG00000106462.6 | enhancer of zeste 2 polycomb repressive complex 2 subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EZH2 | hg19_v2_chr7_-_148581251_148581347 | -0.82 | 1.8e-01 | Click! |
Activity profile of EZH2 motif
Sorted Z-values of EZH2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_51152702 | 2.05 |
ENST00000425202.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr5_+_175298487 | 2.04 |
ENST00000393745.3
|
CPLX2
|
complexin 2 |
| chr5_+_175298573 | 1.82 |
ENST00000512824.1
|
CPLX2
|
complexin 2 |
| chr5_+_175298674 | 1.57 |
ENST00000514150.1
|
CPLX2
|
complexin 2 |
| chr19_-_56056888 | 0.81 |
ENST00000592464.1
ENST00000420723.3 |
SBK3
|
SH3 domain binding kinase family, member 3 |
| chr15_+_91416092 | 0.77 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr10_-_75168071 | 0.77 |
ENST00000394847.3
|
ANXA7
|
annexin A7 |
| chr9_+_131902283 | 0.75 |
ENST00000436883.1
ENST00000414510.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr11_-_44331679 | 0.74 |
ENST00000329255.3
|
ALX4
|
ALX homeobox 4 |
| chrY_+_15418467 | 0.73 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr16_+_30709530 | 0.71 |
ENST00000411466.2
|
SRCAP
|
Snf2-related CREBBP activator protein |
| chr17_+_7123207 | 0.69 |
ENST00000584103.1
ENST00000579886.2 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr5_+_149865377 | 0.68 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr5_+_92919043 | 0.58 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr11_-_66056596 | 0.55 |
ENST00000471387.2
ENST00000359461.6 ENST00000376901.4 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr10_-_81932771 | 0.52 |
ENST00000437799.1
|
ANXA11
|
annexin A11 |
| chr9_-_14322319 | 0.52 |
ENST00000606230.1
|
NFIB
|
nuclear factor I/B |
| chr17_+_75316336 | 0.48 |
ENST00000591934.1
|
SEPT9
|
septin 9 |
| chr8_+_22428457 | 0.45 |
ENST00000517962.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr12_+_7022909 | 0.44 |
ENST00000537688.1
|
ENO2
|
enolase 2 (gamma, neuronal) |
| chr2_+_121493717 | 0.44 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr5_+_102200948 | 0.44 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr17_+_55183261 | 0.43 |
ENST00000576295.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr9_+_131902346 | 0.42 |
ENST00000432124.1
ENST00000435305.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr5_+_158527630 | 0.42 |
ENST00000523301.1
|
RP11-175K6.1
|
RP11-175K6.1 |
| chr19_+_1261106 | 0.42 |
ENST00000588411.1
|
CIRBP
|
cold inducible RNA binding protein |
| chr14_-_23284703 | 0.41 |
ENST00000555911.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr14_+_77244349 | 0.40 |
ENST00000554743.1
|
VASH1
|
vasohibin 1 |
| chrX_+_152990302 | 0.40 |
ENST00000218104.3
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr16_+_30662085 | 0.40 |
ENST00000569864.1
|
PRR14
|
proline rich 14 |
| chr7_+_13141010 | 0.39 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr3_-_157824292 | 0.38 |
ENST00000483851.2
|
SHOX2
|
short stature homeobox 2 |
| chr16_-_1020954 | 0.38 |
ENST00000543238.1
ENST00000539379.1 ENST00000399843.2 ENST00000262301.11 |
LMF1
|
lipase maturation factor 1 |
| chr11_+_57559005 | 0.37 |
ENST00000534647.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr2_+_210288760 | 0.37 |
ENST00000199940.6
|
MAP2
|
microtubule-associated protein 2 |
| chr3_-_120400960 | 0.36 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr6_-_31697255 | 0.36 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr3_-_129035120 | 0.36 |
ENST00000333762.4
|
H1FX
|
H1 histone family, member X |
| chr2_+_220462560 | 0.36 |
ENST00000456909.1
ENST00000295641.10 |
STK11IP
|
serine/threonine kinase 11 interacting protein |
| chr14_-_23285069 | 0.36 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr6_-_113754604 | 0.35 |
ENST00000421737.1
|
RP1-124C6.1
|
RP1-124C6.1 |
| chr4_-_159094194 | 0.35 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr13_-_45048386 | 0.35 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr6_+_30848771 | 0.35 |
ENST00000503180.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr17_-_7387524 | 0.35 |
ENST00000311403.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chr2_-_220436248 | 0.35 |
ENST00000265318.4
|
OBSL1
|
obscurin-like 1 |
| chr16_+_29911864 | 0.35 |
ENST00000308748.5
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr5_+_175223313 | 0.34 |
ENST00000359546.4
|
CPLX2
|
complexin 2 |
| chr22_-_39268192 | 0.34 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chrX_+_100878112 | 0.33 |
ENST00000491568.2
ENST00000479298.1 |
ARMCX3
|
armadillo repeat containing, X-linked 3 |
| chr17_-_46806540 | 0.33 |
ENST00000290295.7
|
HOXB13
|
homeobox B13 |
| chr17_-_47045949 | 0.33 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr16_-_89785777 | 0.33 |
ENST00000561976.1
|
VPS9D1
|
VPS9 domain containing 1 |
| chr16_-_19725899 | 0.33 |
ENST00000567367.1
|
KNOP1
|
lysine-rich nucleolar protein 1 |
| chr5_-_158526756 | 0.32 |
ENST00000313708.6
ENST00000517373.1 |
EBF1
|
early B-cell factor 1 |
| chr19_-_46405861 | 0.32 |
ENST00000322217.5
|
MYPOP
|
Myb-related transcription factor, partner of profilin |
| chr7_-_100881109 | 0.32 |
ENST00000308344.5
|
CLDN15
|
claudin 15 |
| chr13_-_108518986 | 0.32 |
ENST00000375915.2
|
FAM155A
|
family with sequence similarity 155, member A |
| chr6_+_30848829 | 0.32 |
ENST00000508317.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr17_-_46724186 | 0.32 |
ENST00000433510.2
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr10_-_77161533 | 0.31 |
ENST00000535216.1
|
ZNF503
|
zinc finger protein 503 |
| chr6_+_35744367 | 0.31 |
ENST00000360454.2
ENST00000403376.3 |
CLPSL2
|
colipase-like 2 |
| chr19_-_36505098 | 0.31 |
ENST00000252984.7
ENST00000486389.1 ENST00000378875.3 ENST00000485128.1 |
ALKBH6
|
alkB, alkylation repair homolog 6 (E. coli) |
| chr5_-_141703713 | 0.30 |
ENST00000511815.1
|
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr17_+_75276643 | 0.30 |
ENST00000589070.1
|
SEPT9
|
septin 9 |
| chr3_-_187455680 | 0.30 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr12_-_6483969 | 0.30 |
ENST00000396966.2
|
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chr2_+_66662249 | 0.29 |
ENST00000560281.2
|
MEIS1
|
Meis homeobox 1 |
| chr1_+_155023757 | 0.29 |
ENST00000356955.2
ENST00000449910.2 ENST00000359280.4 ENST00000360674.4 ENST00000368412.3 ENST00000355956.2 ENST00000368410.2 ENST00000271836.6 ENST00000368413.1 ENST00000531455.1 ENST00000447332.3 |
ADAM15
|
ADAM metallopeptidase domain 15 |
| chr17_-_45918539 | 0.29 |
ENST00000584123.1
ENST00000578323.1 ENST00000407215.3 ENST00000290216.9 |
SCRN2
|
secernin 2 |
| chr9_+_127539425 | 0.29 |
ENST00000331715.9
|
OLFML2A
|
olfactomedin-like 2A |
| chr1_+_225600404 | 0.28 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr2_-_118943930 | 0.28 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr12_+_6930703 | 0.28 |
ENST00000311268.3
|
GPR162
|
G protein-coupled receptor 162 |
| chr2_-_190044480 | 0.28 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr15_-_88799948 | 0.28 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr3_-_112360116 | 0.27 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr18_+_72166564 | 0.27 |
ENST00000583216.1
ENST00000581912.1 ENST00000582589.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr19_+_41903709 | 0.27 |
ENST00000542943.1
ENST00000457836.2 |
BCKDHA
|
branched chain keto acid dehydrogenase E1, alpha polypeptide |
| chr22_-_39268308 | 0.27 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr2_+_28615669 | 0.27 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr12_-_6715808 | 0.27 |
ENST00000545584.1
|
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr17_-_8027402 | 0.27 |
ENST00000541682.2
ENST00000317814.4 ENST00000577735.1 |
HES7
|
hes family bHLH transcription factor 7 |
| chr12_+_6930813 | 0.27 |
ENST00000428545.2
|
GPR162
|
G protein-coupled receptor 162 |
| chr15_+_81293254 | 0.27 |
ENST00000267984.2
|
MESDC1
|
mesoderm development candidate 1 |
| chr16_+_31085714 | 0.27 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr1_+_157963063 | 0.27 |
ENST00000360089.4
ENST00000368173.3 ENST00000392272.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr2_+_217363559 | 0.27 |
ENST00000600880.1
ENST00000446558.1 |
RPL37A
|
ribosomal protein L37a |
| chr19_-_12992244 | 0.27 |
ENST00000538460.1
|
DNASE2
|
deoxyribonuclease II, lysosomal |
| chr17_+_21729899 | 0.26 |
ENST00000583708.1
|
UBBP4
|
ubiquitin B pseudogene 4 |
| chr4_-_153457197 | 0.26 |
ENST00000281708.4
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr16_-_29934558 | 0.26 |
ENST00000568995.1
ENST00000566413.1 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr7_+_140396465 | 0.26 |
ENST00000476279.1
ENST00000247866.4 ENST00000461457.1 ENST00000465506.1 ENST00000204307.5 ENST00000464566.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr1_+_20878932 | 0.26 |
ENST00000332947.4
|
FAM43B
|
family with sequence similarity 43, member B |
| chr17_+_4846101 | 0.26 |
ENST00000576965.1
|
RNF167
|
ring finger protein 167 |
| chr6_-_43484621 | 0.25 |
ENST00000506469.1
ENST00000503972.1 |
YIPF3
|
Yip1 domain family, member 3 |
| chr6_-_168397757 | 0.25 |
ENST00000456585.1
ENST00000414364.1 |
KIF25-AS1
|
KIF25 antisense RNA 1 |
| chrX_+_70316005 | 0.25 |
ENST00000374259.3
|
FOXO4
|
forkhead box O4 |
| chr6_+_32132360 | 0.25 |
ENST00000333845.6
ENST00000395512.1 ENST00000432129.1 |
EGFL8
|
EGF-like-domain, multiple 8 |
| chr17_+_42148225 | 0.25 |
ENST00000591696.1
|
G6PC3
|
glucose 6 phosphatase, catalytic, 3 |
| chr8_+_22857048 | 0.25 |
ENST00000251822.6
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr1_-_150208320 | 0.25 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr19_-_40440533 | 0.25 |
ENST00000221347.6
|
FCGBP
|
Fc fragment of IgG binding protein |
| chr16_+_31483451 | 0.25 |
ENST00000565360.1
ENST00000361773.3 |
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr12_-_56652111 | 0.25 |
ENST00000267116.7
|
ANKRD52
|
ankyrin repeat domain 52 |
| chr12_-_116714564 | 0.24 |
ENST00000548743.1
|
MED13L
|
mediator complex subunit 13-like |
| chr7_-_100076873 | 0.24 |
ENST00000300181.2
|
TSC22D4
|
TSC22 domain family, member 4 |
| chr3_-_15563229 | 0.24 |
ENST00000383786.5
ENST00000383787.2 ENST00000383785.2 ENST00000383788.5 ENST00000603808.1 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr22_-_29663954 | 0.24 |
ENST00000216085.7
|
RHBDD3
|
rhomboid domain containing 3 |
| chr8_+_22423168 | 0.24 |
ENST00000518912.1
ENST00000428103.1 |
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr5_+_137673200 | 0.24 |
ENST00000434981.2
|
FAM53C
|
family with sequence similarity 53, member C |
| chr6_-_37665751 | 0.24 |
ENST00000297153.7
ENST00000434837.3 |
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr11_-_117698765 | 0.24 |
ENST00000532119.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr10_-_77161650 | 0.24 |
ENST00000372524.4
|
ZNF503
|
zinc finger protein 503 |
| chr4_-_80994471 | 0.24 |
ENST00000295465.4
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr5_+_59783941 | 0.24 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr3_-_197300194 | 0.24 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr17_-_40575535 | 0.24 |
ENST00000357037.5
|
PTRF
|
polymerase I and transcript release factor |
| chr16_-_29875057 | 0.24 |
ENST00000219789.6
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr12_+_113376249 | 0.24 |
ENST00000551007.1
ENST00000548514.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr9_-_34372830 | 0.24 |
ENST00000379142.3
|
KIAA1161
|
KIAA1161 |
| chr14_-_23285011 | 0.23 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr12_+_6930964 | 0.23 |
ENST00000382315.3
|
GPR162
|
G protein-coupled receptor 162 |
| chr17_+_7358889 | 0.23 |
ENST00000575379.1
|
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr12_+_66218212 | 0.22 |
ENST00000393578.3
ENST00000425208.2 ENST00000536545.1 ENST00000354636.3 |
HMGA2
|
high mobility group AT-hook 2 |
| chr16_+_69373471 | 0.22 |
ENST00000569637.2
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr6_-_32143828 | 0.22 |
ENST00000412465.2
ENST00000375107.3 |
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr2_-_216003127 | 0.22 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr15_+_64428529 | 0.22 |
ENST00000560861.1
|
SNX1
|
sorting nexin 1 |
| chr1_+_45274154 | 0.22 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr22_+_24990746 | 0.22 |
ENST00000456869.1
ENST00000411974.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr19_-_54692132 | 0.22 |
ENST00000449249.1
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7 |
| chr18_+_3450161 | 0.22 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr3_+_164924716 | 0.22 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr11_+_64008525 | 0.22 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr10_-_75682535 | 0.22 |
ENST00000409178.1
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr17_+_27895045 | 0.22 |
ENST00000580183.2
ENST00000578749.1 ENST00000582829.2 |
TP53I13
|
tumor protein p53 inducible protein 13 |
| chr2_-_27435634 | 0.22 |
ENST00000430186.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr1_-_153521714 | 0.22 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr9_+_130469257 | 0.22 |
ENST00000373295.2
|
C9orf117
|
chromosome 9 open reading frame 117 |
| chr7_-_100076765 | 0.21 |
ENST00000393991.1
|
TSC22D4
|
TSC22 domain family, member 4 |
| chr11_+_64008443 | 0.21 |
ENST00000309366.4
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr6_+_32936942 | 0.21 |
ENST00000496118.2
|
BRD2
|
bromodomain containing 2 |
| chr16_-_30773372 | 0.21 |
ENST00000545825.1
ENST00000541260.1 |
C16orf93
|
chromosome 16 open reading frame 93 |
| chr12_-_118406777 | 0.21 |
ENST00000339824.5
|
KSR2
|
kinase suppressor of ras 2 |
| chr16_+_31483374 | 0.21 |
ENST00000394863.3
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr17_-_71223839 | 0.21 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr20_-_62168714 | 0.21 |
ENST00000542869.1
|
PTK6
|
protein tyrosine kinase 6 |
| chr1_-_155112883 | 0.20 |
ENST00000368399.1
ENST00000368400.4 ENST00000341298.3 |
DPM3
|
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr16_-_29910853 | 0.20 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr11_-_6255846 | 0.20 |
ENST00000449352.2
|
FAM160A2
|
family with sequence similarity 160, member A2 |
| chr17_-_56494882 | 0.20 |
ENST00000584437.1
|
RNF43
|
ring finger protein 43 |
| chr16_-_2260834 | 0.20 |
ENST00000562360.1
ENST00000566018.1 |
BRICD5
|
BRICHOS domain containing 5 |
| chr9_+_35829208 | 0.20 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr15_+_59903975 | 0.20 |
ENST00000560585.1
ENST00000396065.1 |
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr5_+_158527485 | 0.20 |
ENST00000517335.1
|
RP11-175K6.1
|
RP11-175K6.1 |
| chr3_+_50654821 | 0.20 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr8_+_22438009 | 0.20 |
ENST00000409417.1
|
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr14_-_61124977 | 0.20 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr1_-_153521597 | 0.20 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr17_+_67498396 | 0.20 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr1_+_145727681 | 0.20 |
ENST00000417171.1
ENST00000451928.2 |
PDZK1
|
PDZ domain containing 1 |
| chr16_-_1020849 | 0.20 |
ENST00000568897.1
|
LMF1
|
lipase maturation factor 1 |
| chr10_-_103454876 | 0.19 |
ENST00000331272.7
|
FBXW4
|
F-box and WD repeat domain containing 4 |
| chr1_-_209824643 | 0.19 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr17_-_46667594 | 0.19 |
ENST00000476342.1
ENST00000460160.1 ENST00000472863.1 |
HOXB3
|
homeobox B3 |
| chr10_+_71389983 | 0.19 |
ENST00000373279.4
|
C10orf35
|
chromosome 10 open reading frame 35 |
| chr15_+_90544532 | 0.19 |
ENST00000268154.4
|
ZNF710
|
zinc finger protein 710 |
| chr19_+_51728316 | 0.19 |
ENST00000436584.2
ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33
|
CD33 molecule |
| chr9_-_34628104 | 0.19 |
ENST00000378909.2
|
ARID3C
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr17_+_42148097 | 0.19 |
ENST00000269097.4
|
G6PC3
|
glucose 6 phosphatase, catalytic, 3 |
| chr2_-_220435963 | 0.19 |
ENST00000373876.1
ENST00000404537.1 ENST00000603926.1 ENST00000373873.4 ENST00000289656.3 |
OBSL1
|
obscurin-like 1 |
| chr1_+_18807424 | 0.19 |
ENST00000400664.1
|
KLHDC7A
|
kelch domain containing 7A |
| chr11_-_62414070 | 0.19 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr16_+_30662050 | 0.19 |
ENST00000568754.1
|
PRR14
|
proline rich 14 |
| chr11_-_17410869 | 0.19 |
ENST00000528731.1
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11 |
| chr17_-_56494908 | 0.19 |
ENST00000577716.1
|
RNF43
|
ring finger protein 43 |
| chr15_-_59041954 | 0.19 |
ENST00000439637.1
ENST00000558004.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr1_-_154934200 | 0.19 |
ENST00000368457.2
|
PYGO2
|
pygopus family PHD finger 2 |
| chr6_+_32146268 | 0.19 |
ENST00000427134.2
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chrX_+_99899180 | 0.19 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr19_-_51289436 | 0.18 |
ENST00000562076.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr1_-_154943212 | 0.18 |
ENST00000368445.5
ENST00000448116.2 ENST00000368449.4 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr10_+_1102303 | 0.18 |
ENST00000381329.1
|
WDR37
|
WD repeat domain 37 |
| chr2_-_131099897 | 0.18 |
ENST00000409127.1
ENST00000437688.2 ENST00000259229.2 |
CCDC115
|
coiled-coil domain containing 115 |
| chr15_-_65117807 | 0.18 |
ENST00000559239.1
ENST00000268043.4 ENST00000333425.6 |
PIF1
|
PIF1 5'-to-3' DNA helicase |
| chr20_-_62168672 | 0.18 |
ENST00000217185.2
|
PTK6
|
protein tyrosine kinase 6 |
| chr7_-_100881041 | 0.18 |
ENST00000412417.1
ENST00000414035.1 |
CLDN15
|
claudin 15 |
| chr4_+_102268904 | 0.18 |
ENST00000527564.1
ENST00000529296.1 |
AP001816.1
|
Uncharacterized protein |
| chr14_+_50234827 | 0.18 |
ENST00000554589.1
ENST00000557247.1 |
KLHDC2
|
kelch domain containing 2 |
| chr17_+_7338737 | 0.18 |
ENST00000323206.1
ENST00000396568.1 |
TMEM102
|
transmembrane protein 102 |
| chr1_+_117544366 | 0.18 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr16_-_29874211 | 0.18 |
ENST00000563415.1
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr9_+_140500087 | 0.18 |
ENST00000371421.4
|
ARRDC1
|
arrestin domain containing 1 |
| chr14_+_23352374 | 0.18 |
ENST00000267396.4
ENST00000536884.1 |
REM2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr6_-_79944336 | 0.18 |
ENST00000344726.5
ENST00000275036.7 |
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr15_+_43885799 | 0.17 |
ENST00000449946.1
ENST00000417289.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr8_+_22423219 | 0.17 |
ENST00000523965.1
ENST00000521554.1 |
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr6_-_43484718 | 0.17 |
ENST00000372422.2
|
YIPF3
|
Yip1 domain family, member 3 |
| chr22_+_29664241 | 0.17 |
ENST00000436425.1
ENST00000447973.1 |
EWSR1
|
EWS RNA-binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EZH2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.2 | 5.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 0.4 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.5 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 1.0 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.4 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.3 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.3 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.4 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.3 | GO:0052026 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.1 | 0.2 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 0.2 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.1 | 0.1 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.1 | 0.6 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.1 | 0.4 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.1 | 0.3 | GO:0021623 | optic cup structural organization(GO:0003409) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.3 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.1 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 0.5 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.4 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.1 | 0.5 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.2 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.1 | 0.2 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.7 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.2 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.7 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.3 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.3 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:2001302 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.3 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.4 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.4 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.4 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.6 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.1 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.3 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.0 | 0.1 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.9 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.3 | GO:0006007 | glucose catabolic process(GO:0006007) NADH metabolic process(GO:0006734) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0052362 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.2 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.7 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.2 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.1 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0002605 | negative regulation of dendritic cell antigen processing and presentation(GO:0002605) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.2 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.3 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.3 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.3 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.2 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.2 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.2 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.2 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.0 | 0.1 | GO:2000481 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 | 0.1 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) renal artery morphogenesis(GO:0061441) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.7 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.1 | GO:0043353 | slow-twitch skeletal muscle fiber contraction(GO:0031444) enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0060482 | lung goblet cell differentiation(GO:0060480) lobar bronchus epithelium development(GO:0060481) lobar bronchus development(GO:0060482) |
| 0.0 | 0.6 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0001079 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.1 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.0 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 0.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 0.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.4 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.0 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.1 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.5 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.5 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.3 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.1 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.2 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.1 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.8 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.4 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.0 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.4 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.8 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 5.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.7 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.1 | 0.3 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.4 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.1 | 0.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.5 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.2 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 0.2 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 0.8 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.1 | 0.4 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.2 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) |
| 0.1 | 0.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.2 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.9 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0005018 | platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 0.0 | 0.2 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.2 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.1 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.3 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.2 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0086040 | sodium:proton antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086040) |
| 0.0 | 0.4 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.1 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 1.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.3 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.0 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.0 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.6 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.5 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |