Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
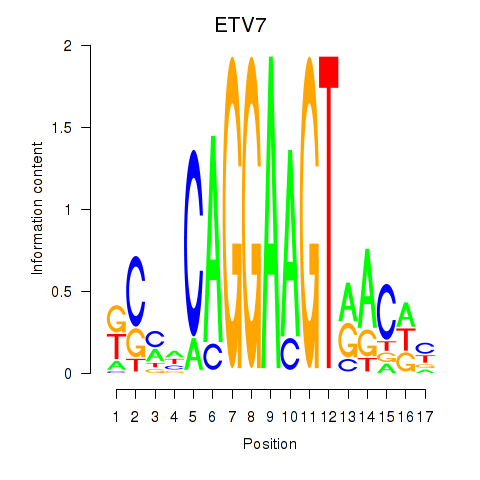
Results for ETV7
Z-value: 1.07
Transcription factors associated with ETV7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV7
|
ENSG00000010030.9 | ETS variant transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV7 | hg19_v2_chr6_-_36355486_36355497 | -0.84 | 1.6e-01 | Click! |
Activity profile of ETV7 motif
Sorted Z-values of ETV7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_139249228 | 0.78 |
ENST00000392944.1
|
GPSM1
|
G-protein signaling modulator 1 |
| chr19_-_53426700 | 0.52 |
ENST00000596623.1
|
ZNF888
|
zinc finger protein 888 |
| chr6_-_33385854 | 0.49 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr19_+_55996565 | 0.49 |
ENST00000587400.1
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr16_+_77225071 | 0.49 |
ENST00000439557.2
ENST00000545553.1 |
MON1B
|
MON1 secretory trafficking family member B |
| chr9_+_140100113 | 0.47 |
ENST00000371521.4
ENST00000344894.5 ENST00000427047.2 ENST00000458322.2 |
NDOR1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr11_-_9336117 | 0.43 |
ENST00000527813.1
ENST00000533723.1 |
TMEM41B
|
transmembrane protein 41B |
| chr14_+_55590646 | 0.43 |
ENST00000553493.1
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr20_+_55926625 | 0.43 |
ENST00000452119.1
|
RAE1
|
ribonucleic acid export 1 |
| chr17_-_79791118 | 0.42 |
ENST00000576431.1
ENST00000575061.1 ENST00000455127.2 ENST00000572645.1 ENST00000538396.1 ENST00000573478.1 |
FAM195B
|
family with sequence similarity 195, member B |
| chr19_-_51611623 | 0.41 |
ENST00000421832.2
|
CTU1
|
cytosolic thiouridylase subunit 1 |
| chr19_+_18530146 | 0.41 |
ENST00000348495.6
ENST00000270061.7 |
SSBP4
|
single stranded DNA binding protein 4 |
| chr5_+_61708488 | 0.40 |
ENST00000505902.1
|
IPO11
|
importin 11 |
| chr19_+_18529674 | 0.39 |
ENST00000597724.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr18_+_44812072 | 0.38 |
ENST00000598649.1
ENST00000586905.2 |
CTD-2130O13.1
|
CTD-2130O13.1 |
| chr15_-_55488817 | 0.38 |
ENST00000569386.1
|
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr16_+_1832902 | 0.38 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr6_-_26235206 | 0.37 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr11_+_705193 | 0.36 |
ENST00000527199.1
|
EPS8L2
|
EPS8-like 2 |
| chr3_+_19988736 | 0.36 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr11_+_2920951 | 0.36 |
ENST00000347936.2
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr17_+_40688190 | 0.35 |
ENST00000225927.2
|
NAGLU
|
N-acetylglucosaminidase, alpha |
| chr19_+_36236491 | 0.35 |
ENST00000591949.1
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr12_+_123459127 | 0.34 |
ENST00000397389.2
ENST00000538755.1 ENST00000536150.1 ENST00000545056.1 ENST00000545612.1 ENST00000538628.1 ENST00000545317.1 |
OGFOD2
|
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr16_-_4401284 | 0.34 |
ENST00000318059.3
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr15_+_90735145 | 0.33 |
ENST00000559792.1
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr1_+_156698743 | 0.32 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr19_+_18530184 | 0.32 |
ENST00000601357.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr1_+_156698234 | 0.32 |
ENST00000368218.4
ENST00000368216.4 |
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr14_-_94595993 | 0.30 |
ENST00000238609.3
|
IFI27L2
|
interferon, alpha-inducible protein 27-like 2 |
| chr6_-_43484621 | 0.30 |
ENST00000506469.1
ENST00000503972.1 |
YIPF3
|
Yip1 domain family, member 3 |
| chr19_-_3500635 | 0.29 |
ENST00000250937.3
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase |
| chr22_+_27068704 | 0.29 |
ENST00000444388.1
ENST00000450963.1 ENST00000449017.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr19_+_36236514 | 0.29 |
ENST00000222266.2
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr7_+_2685164 | 0.29 |
ENST00000400376.2
|
TTYH3
|
tweety family member 3 |
| chr19_-_50370799 | 0.29 |
ENST00000600910.1
ENST00000322344.3 ENST00000600573.1 |
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr19_+_50145328 | 0.28 |
ENST00000360565.3
|
SCAF1
|
SR-related CTD-associated factor 1 |
| chr13_-_45048386 | 0.28 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr9_+_131085095 | 0.28 |
ENST00000372875.3
|
COQ4
|
coenzyme Q4 |
| chr20_-_34542548 | 0.28 |
ENST00000305978.2
|
SCAND1
|
SCAN domain containing 1 |
| chr12_+_110011571 | 0.28 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr4_-_860950 | 0.27 |
ENST00000511980.1
ENST00000510799.1 |
GAK
|
cyclin G associated kinase |
| chr11_-_62359027 | 0.27 |
ENST00000494385.1
ENST00000308436.7 |
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr19_-_41222775 | 0.27 |
ENST00000324464.3
ENST00000450541.1 ENST00000594720.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr11_-_417308 | 0.26 |
ENST00000397632.3
ENST00000382520.2 |
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr17_+_79849782 | 0.26 |
ENST00000392376.3
|
ANAPC11
|
anaphase promoting complex subunit 11 |
| chr9_-_139010696 | 0.26 |
ENST00000418388.1
ENST00000561457.1 |
C9orf69
|
chromosome 9 open reading frame 69 |
| chr6_-_33267101 | 0.26 |
ENST00000497454.1
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr17_+_78193443 | 0.25 |
ENST00000577155.1
|
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr14_-_23504087 | 0.25 |
ENST00000493471.2
ENST00000460922.2 |
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr11_-_805224 | 0.25 |
ENST00000347755.5
|
PIDD
|
p53-induced death domain protein |
| chr19_+_34287174 | 0.25 |
ENST00000587559.1
ENST00000588637.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr9_-_139891165 | 0.24 |
ENST00000494426.1
|
CLIC3
|
chloride intracellular channel 3 |
| chrX_-_80457385 | 0.24 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr1_+_84767289 | 0.24 |
ENST00000394834.3
ENST00000370669.1 |
SAMD13
|
sterile alpha motif domain containing 13 |
| chr20_+_55926566 | 0.23 |
ENST00000411894.1
ENST00000429339.1 |
RAE1
|
ribonucleic acid export 1 |
| chr9_-_140100068 | 0.23 |
ENST00000537254.1
ENST00000343666.5 |
TMEM203
|
transmembrane protein 203 |
| chr14_+_24563262 | 0.23 |
ENST00000559250.1
ENST00000216780.4 ENST00000560736.1 ENST00000396973.4 ENST00000559837.1 |
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr3_+_149191723 | 0.23 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr17_-_80009650 | 0.23 |
ENST00000310496.4
|
RFNG
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr16_-_30366672 | 0.23 |
ENST00000305596.3
|
CD2BP2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr17_+_80193644 | 0.22 |
ENST00000582946.1
|
SLC16A3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr14_-_23504337 | 0.22 |
ENST00000361611.6
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr11_-_62358972 | 0.22 |
ENST00000278279.3
|
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr1_-_1677358 | 0.22 |
ENST00000355439.2
ENST00000400924.1 ENST00000246421.4 |
SLC35E2
|
solute carrier family 35, member E2 |
| chr5_-_180237082 | 0.22 |
ENST00000506889.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr19_-_8579030 | 0.22 |
ENST00000255616.8
ENST00000393927.4 |
ZNF414
|
zinc finger protein 414 |
| chr16_+_1543337 | 0.22 |
ENST00000262319.6
|
TELO2
|
telomere maintenance 2 |
| chr12_+_34175398 | 0.21 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr2_-_220041617 | 0.21 |
ENST00000451647.1
ENST00000360507.5 |
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr7_-_5553369 | 0.21 |
ENST00000453700.3
ENST00000382368.3 |
FBXL18
|
F-box and leucine-rich repeat protein 18 |
| chr12_+_94071129 | 0.21 |
ENST00000552983.1
ENST00000332896.3 ENST00000552033.1 ENST00000548483.1 |
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr16_+_838614 | 0.21 |
ENST00000262315.9
ENST00000455171.2 |
CHTF18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) |
| chr12_+_51633061 | 0.21 |
ENST00000551313.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr5_-_180236811 | 0.21 |
ENST00000446023.2
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr6_+_31515337 | 0.21 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr13_+_28527647 | 0.21 |
ENST00000567234.1
|
LINC00543
|
long intergenic non-protein coding RNA 543 |
| chr20_+_62367989 | 0.21 |
ENST00000309546.3
|
LIME1
|
Lck interacting transmembrane adaptor 1 |
| chr5_-_158757895 | 0.20 |
ENST00000231228.2
|
IL12B
|
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) |
| chr6_-_43484718 | 0.20 |
ENST00000372422.2
|
YIPF3
|
Yip1 domain family, member 3 |
| chr17_+_79849872 | 0.20 |
ENST00000584197.1
ENST00000583839.1 |
ANAPC11
|
anaphase promoting complex subunit 11 |
| chr7_-_1609591 | 0.20 |
ENST00000288607.2
ENST00000404674.3 |
PSMG3
|
proteasome (prosome, macropain) assembly chaperone 3 |
| chr19_+_11466062 | 0.20 |
ENST00000251473.5
ENST00000591329.1 ENST00000586380.1 |
DKFZP761J1410
|
Lipid phosphate phosphatase-related protein type 2 |
| chr1_+_153963227 | 0.20 |
ENST00000368567.4
ENST00000392558.4 |
RPS27
|
ribosomal protein S27 |
| chr19_-_913160 | 0.20 |
ENST00000361574.5
ENST00000587975.1 |
R3HDM4
|
R3H domain containing 4 |
| chr5_+_180650271 | 0.19 |
ENST00000351937.5
ENST00000315073.5 |
TRIM41
|
tripartite motif containing 41 |
| chr16_-_71323617 | 0.19 |
ENST00000563876.1
|
CMTR2
|
cap methyltransferase 2 |
| chr11_-_64885111 | 0.19 |
ENST00000528598.1
ENST00000310597.4 |
ZNHIT2
|
zinc finger, HIT-type containing 2 |
| chr1_-_249120832 | 0.19 |
ENST00000366472.5
|
SH3BP5L
|
SH3-binding domain protein 5-like |
| chr16_+_30205225 | 0.19 |
ENST00000345535.4
ENST00000251303.6 |
SLX1A
|
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae) |
| chr19_+_3762645 | 0.19 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr1_+_156698708 | 0.19 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr11_-_67771513 | 0.19 |
ENST00000227471.2
|
UNC93B1
|
unc-93 homolog B1 (C. elegans) |
| chr18_+_3411595 | 0.18 |
ENST00000552383.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr17_+_49243639 | 0.18 |
ENST00000512737.1
ENST00000503064.1 |
NME1-NME2
|
NME1-NME2 readthrough |
| chr2_+_241544834 | 0.18 |
ENST00000319838.5
ENST00000403859.1 ENST00000438013.2 |
GPR35
|
G protein-coupled receptor 35 |
| chr11_-_417388 | 0.18 |
ENST00000332725.3
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr7_-_1543981 | 0.18 |
ENST00000404767.3
|
INTS1
|
integrator complex subunit 1 |
| chr7_+_1609694 | 0.18 |
ENST00000437621.2
ENST00000457484.2 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr2_-_220041860 | 0.18 |
ENST00000453038.1
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr2_-_200715573 | 0.18 |
ENST00000420922.2
|
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chr6_-_166755995 | 0.17 |
ENST00000361731.3
|
SFT2D1
|
SFT2 domain containing 1 |
| chr7_+_1609765 | 0.17 |
ENST00000437964.1
ENST00000533935.1 ENST00000532358.1 ENST00000524978.1 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr17_+_19281034 | 0.17 |
ENST00000308406.5
ENST00000299612.7 |
MAPK7
|
mitogen-activated protein kinase 7 |
| chr6_-_34855773 | 0.17 |
ENST00000420584.2
ENST00000361288.4 |
TAF11
|
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa |
| chr1_-_1655713 | 0.17 |
ENST00000401096.2
ENST00000404249.3 ENST00000357760.2 ENST00000358779.5 ENST00000378633.1 ENST00000378635.3 |
CDK11A
|
cyclin-dependent kinase 11A |
| chr2_+_87808725 | 0.17 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr19_-_12792704 | 0.17 |
ENST00000210060.7
|
DHPS
|
deoxyhypusine synthase |
| chr20_+_35918035 | 0.17 |
ENST00000373606.3
ENST00000397156.3 ENST00000397150.1 ENST00000397152.3 |
MANBAL
|
mannosidase, beta A, lysosomal-like |
| chr12_+_105724613 | 0.17 |
ENST00000549934.2
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr16_+_29465822 | 0.17 |
ENST00000330181.5
ENST00000351581.4 |
SLX1B
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr19_+_13080296 | 0.17 |
ENST00000317060.2
|
DAND5
|
DAN domain family member 5, BMP antagonist |
| chr1_+_27648648 | 0.17 |
ENST00000374076.4
|
TMEM222
|
transmembrane protein 222 |
| chr17_-_7155775 | 0.17 |
ENST00000571409.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr17_+_79849523 | 0.17 |
ENST00000572639.1
ENST00000579978.1 ENST00000344877.5 ENST00000582222.1 ENST00000577425.1 ENST00000571024.2 ENST00000574924.2 ENST00000572851.2 ENST00000357385.3 ENST00000584314.1 ENST00000571874.2 ENST00000578550.1 ENST00000577747.1 ENST00000579133.1 |
ANAPC11
|
anaphase promoting complex subunit 11 |
| chr19_+_55385731 | 0.17 |
ENST00000469767.1
ENST00000355524.3 ENST00000391725.3 ENST00000345937.4 ENST00000353758.4 ENST00000359272.4 ENST00000391723.3 ENST00000391724.3 |
FCAR
|
Fc fragment of IgA, receptor for |
| chr11_+_65337901 | 0.17 |
ENST00000309328.3
ENST00000531405.1 ENST00000527920.1 ENST00000526877.1 ENST00000533115.1 ENST00000526433.1 |
SSSCA1
|
Sjogren syndrome/scleroderma autoantigen 1 |
| chr19_+_55996543 | 0.17 |
ENST00000591590.1
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr16_-_89785777 | 0.17 |
ENST00000561976.1
|
VPS9D1
|
VPS9 domain containing 1 |
| chr3_+_9932238 | 0.17 |
ENST00000307768.4
|
JAGN1
|
jagunal homolog 1 (Drosophila) |
| chr1_-_155225178 | 0.16 |
ENST00000361361.2
|
FAM189B
|
family with sequence similarity 189, member B |
| chr9_+_131084815 | 0.16 |
ENST00000300452.3
ENST00000609948.1 |
COQ4
|
coenzyme Q4 |
| chr22_+_27068766 | 0.16 |
ENST00000435162.1
ENST00000437071.1 ENST00000440816.1 ENST00000421253.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr2_+_217363826 | 0.16 |
ENST00000441179.2
|
RPL37A
|
ribosomal protein L37a |
| chr1_+_3689325 | 0.16 |
ENST00000444870.2
ENST00000452264.1 |
SMIM1
|
small integral membrane protein 1 (Vel blood group) |
| chr17_-_79269067 | 0.16 |
ENST00000288439.5
ENST00000374759.3 |
SLC38A10
|
solute carrier family 38, member 10 |
| chr20_-_2451395 | 0.16 |
ENST00000339610.6
ENST00000381342.2 ENST00000438552.2 |
SNRPB
|
small nuclear ribonucleoprotein polypeptides B and B1 |
| chr15_-_65117807 | 0.16 |
ENST00000559239.1
ENST00000268043.4 ENST00000333425.6 |
PIF1
|
PIF1 5'-to-3' DNA helicase |
| chr16_+_89627061 | 0.16 |
ENST00000311528.5
ENST00000563270.1 ENST00000567815.1 ENST00000452368.3 ENST00000467736.1 ENST00000393099.3 |
RPL13
|
ribosomal protein L13 |
| chr14_-_21567009 | 0.16 |
ENST00000556174.1
ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219
|
zinc finger protein 219 |
| chr16_-_734318 | 0.16 |
ENST00000609261.1
ENST00000562111.1 ENST00000562824.1 ENST00000412368.2 ENST00000293882.4 ENST00000454700.1 |
JMJD8
|
jumonji domain containing 8 |
| chr1_+_24742264 | 0.16 |
ENST00000374399.4
ENST00000003912.3 ENST00000358028.4 ENST00000339255.2 |
NIPAL3
|
NIPA-like domain containing 3 |
| chr3_+_160117062 | 0.16 |
ENST00000497311.1
|
SMC4
|
structural maintenance of chromosomes 4 |
| chr1_+_156024552 | 0.16 |
ENST00000368304.5
ENST00000368302.3 |
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr11_+_64008443 | 0.16 |
ENST00000309366.4
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr16_+_30953696 | 0.16 |
ENST00000566320.2
ENST00000565939.1 |
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr20_+_60758075 | 0.15 |
ENST00000536470.1
ENST00000436421.2 ENST00000370823.3 ENST00000448254.1 |
MTG2
|
mitochondrial ribosome-associated GTPase 2 |
| chr17_-_33288522 | 0.15 |
ENST00000314144.5
|
CCT6B
|
chaperonin containing TCP1, subunit 6B (zeta 2) |
| chr1_+_154300217 | 0.15 |
ENST00000368489.3
|
ATP8B2
|
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr5_-_180237445 | 0.15 |
ENST00000393340.3
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr19_+_42772659 | 0.15 |
ENST00000572681.2
|
CIC
|
capicua transcriptional repressor |
| chr12_-_58329819 | 0.15 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr1_-_155112883 | 0.15 |
ENST00000368399.1
ENST00000368400.4 ENST00000341298.3 |
DPM3
|
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr14_+_24899141 | 0.15 |
ENST00000556842.1
ENST00000553935.1 |
KHNYN
|
KH and NYN domain containing |
| chr17_+_74722912 | 0.15 |
ENST00000589977.1
ENST00000591571.1 ENST00000592849.1 ENST00000586738.1 ENST00000588783.1 ENST00000588563.1 ENST00000586752.1 ENST00000588302.1 ENST00000590964.1 ENST00000341249.6 ENST00000588822.1 |
METTL23
|
methyltransferase like 23 |
| chr11_-_60929074 | 0.15 |
ENST00000301765.5
|
VPS37C
|
vacuolar protein sorting 37 homolog C (S. cerevisiae) |
| chr19_+_51153045 | 0.15 |
ENST00000458538.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr19_-_40971643 | 0.15 |
ENST00000595483.1
|
BLVRB
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr1_-_94344686 | 0.15 |
ENST00000528680.1
|
DNTTIP2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr14_-_23504432 | 0.15 |
ENST00000425762.2
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr5_-_72861175 | 0.14 |
ENST00000504641.1
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr3_-_47950745 | 0.14 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr7_+_100860949 | 0.14 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr2_-_220436248 | 0.14 |
ENST00000265318.4
|
OBSL1
|
obscurin-like 1 |
| chrX_-_7895755 | 0.14 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr22_-_36877371 | 0.14 |
ENST00000403313.1
|
TXN2
|
thioredoxin 2 |
| chr17_-_6917755 | 0.14 |
ENST00000593646.1
|
AC040977.1
|
Uncharacterized protein |
| chr1_-_226111929 | 0.14 |
ENST00000343818.6
ENST00000432920.2 |
PYCR2
RP4-559A3.7
|
pyrroline-5-carboxylate reductase family, member 2 Uncharacterized protein |
| chr10_-_126432821 | 0.14 |
ENST00000280780.6
|
FAM53B
|
family with sequence similarity 53, member B |
| chr1_-_39407467 | 0.14 |
ENST00000540558.1
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr19_-_4302375 | 0.14 |
ENST00000600114.1
ENST00000600349.1 ENST00000595645.1 ENST00000301272.2 |
TMIGD2
|
transmembrane and immunoglobulin domain containing 2 |
| chr7_+_2281843 | 0.13 |
ENST00000356714.1
ENST00000397049.1 |
NUDT1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 1 |
| chr19_+_41222998 | 0.13 |
ENST00000263370.2
|
ITPKC
|
inositol-trisphosphate 3-kinase C |
| chr1_-_32687923 | 0.13 |
ENST00000309777.6
ENST00000344461.3 ENST00000373593.1 ENST00000545122.1 |
TMEM234
|
transmembrane protein 234 |
| chr16_-_4401258 | 0.13 |
ENST00000577031.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr10_-_27444143 | 0.13 |
ENST00000477432.1
|
YME1L1
|
YME1-like 1 ATPase |
| chr9_+_139553306 | 0.13 |
ENST00000371699.1
|
EGFL7
|
EGF-like-domain, multiple 7 |
| chr11_+_62554860 | 0.13 |
ENST00000533861.1
ENST00000333449.4 |
TMEM179B
|
transmembrane protein 179B |
| chr7_-_7680601 | 0.13 |
ENST00000396682.2
|
RPA3
|
replication protein A3, 14kDa |
| chr14_-_24711764 | 0.13 |
ENST00000557921.1
ENST00000558476.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr9_-_91793675 | 0.13 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr17_+_7255208 | 0.13 |
ENST00000333751.3
|
KCTD11
|
potassium channel tetramerization domain containing 11 |
| chr9_-_38069208 | 0.13 |
ENST00000377707.3
ENST00000377700.4 |
SHB
|
Src homology 2 domain containing adaptor protein B |
| chr5_+_140044261 | 0.13 |
ENST00000358337.5
|
WDR55
|
WD repeat domain 55 |
| chr9_-_100000957 | 0.13 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr12_-_7596735 | 0.13 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr22_+_30805086 | 0.13 |
ENST00000439838.1
ENST00000439023.3 |
RP4-539M6.19
|
Uncharacterized protein |
| chr1_+_156024525 | 0.13 |
ENST00000368305.4
|
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr22_-_31688431 | 0.13 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr1_+_43855545 | 0.13 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr10_+_127585118 | 0.13 |
ENST00000449042.2
|
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr2_+_74685413 | 0.13 |
ENST00000233615.2
|
WBP1
|
WW domain binding protein 1 |
| chr8_+_95565947 | 0.13 |
ENST00000523011.1
|
RP11-267M23.4
|
RP11-267M23.4 |
| chr16_-_28503327 | 0.13 |
ENST00000535392.1
ENST00000395653.4 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr19_+_35739897 | 0.13 |
ENST00000605618.1
ENST00000427250.1 ENST00000601623.1 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr8_-_121457608 | 0.12 |
ENST00000306185.3
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr15_+_90895471 | 0.12 |
ENST00000354377.3
ENST00000379090.5 |
ZNF774
|
zinc finger protein 774 |
| chr19_-_12792585 | 0.12 |
ENST00000351660.5
|
DHPS
|
deoxyhypusine synthase |
| chr17_-_73892992 | 0.12 |
ENST00000540128.1
ENST00000269383.3 |
TRIM65
|
tripartite motif containing 65 |
| chr9_-_131085021 | 0.12 |
ENST00000372890.4
|
TRUB2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr19_-_58400148 | 0.12 |
ENST00000595048.1
ENST00000600634.1 ENST00000595295.1 ENST00000596604.1 ENST00000597342.1 ENST00000597807.1 |
ZNF814
|
zinc finger protein 814 |
| chr6_-_31620455 | 0.12 |
ENST00000437771.1
ENST00000404765.2 ENST00000375964.6 ENST00000211379.5 |
BAG6
|
BCL2-associated athanogene 6 |
| chr11_-_62477103 | 0.12 |
ENST00000532818.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr22_-_31688381 | 0.12 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr19_-_57988871 | 0.12 |
ENST00000596831.1
ENST00000601768.1 ENST00000356584.3 ENST00000600175.1 ENST00000425074.3 ENST00000343280.4 ENST00000427512.2 |
AC004076.9
ZNF772
|
Uncharacterized protein zinc finger protein 772 |
| chr17_+_38474489 | 0.12 |
ENST00000394089.2
ENST00000425707.3 |
RARA
|
retinoic acid receptor, alpha |
| chr20_+_1246908 | 0.12 |
ENST00000381873.3
ENST00000381867.1 |
SNPH
|
syntaphilin |
| chr18_+_72166564 | 0.12 |
ENST00000583216.1
ENST00000581912.1 ENST00000582589.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr11_+_66824276 | 0.12 |
ENST00000308831.2
|
RHOD
|
ras homolog family member D |
| chr11_-_67210930 | 0.12 |
ENST00000453768.2
ENST00000545016.1 ENST00000341356.5 |
CORO1B
|
coronin, actin binding protein, 1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.6 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.3 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.1 | 0.3 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.7 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.1 | 0.4 | GO:2000521 | positive regulation of mononuclear cell migration(GO:0071677) negative regulation of immunological synapse formation(GO:2000521) |
| 0.1 | 0.3 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.2 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.1 | 0.3 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.1 | 0.3 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.1 | 0.6 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.4 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.2 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) negative regulation of ERK5 cascade(GO:0070377) |
| 0.1 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.2 | GO:1903939 | regulation of TORC2 signaling(GO:1903939) |
| 0.1 | 0.4 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 0.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.3 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.7 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.2 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.0 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.3 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.2 | GO:2000330 | positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.6 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:0035419 | activation of MAPK activity involved in innate immune response(GO:0035419) regulation of thymocyte migration(GO:2000410) |
| 0.0 | 0.1 | GO:0002583 | regulation of antigen processing and presentation of peptide antigen(GO:0002583) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.1 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.3 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.3 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.0 | 0.2 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.7 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.7 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.6 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.6 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.0 | 0.1 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.2 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.0 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.0 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.2 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.0 | 0.2 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.0 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.0 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.0 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.4 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 0.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.7 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.2 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.3 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.0 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.0 | GO:0098576 | lumenal side of membrane(GO:0098576) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.3 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 0.3 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.1 | 0.3 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.4 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 0.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.3 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.5 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 0.3 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 0.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.2 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.1 | 0.4 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.2 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) |
| 0.1 | 0.2 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.0 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 0.5 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.1 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.4 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.2 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |