Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
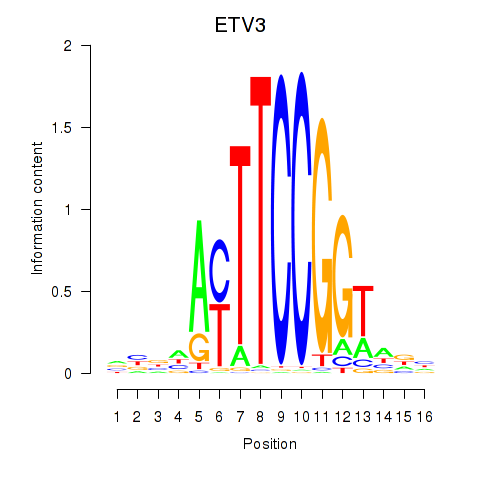
Results for ETV3
Z-value: 0.47
Transcription factors associated with ETV3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV3
|
ENSG00000117036.7 | ETS variant transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV3 | hg19_v2_chr1_-_157108130_157108173 | 0.99 | 5.2e-03 | Click! |
Activity profile of ETV3 motif
Sorted Z-values of ETV3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_71184724 | 0.35 |
ENST00000560604.1
|
THAP10
|
THAP domain containing 10 |
| chr3_-_47324008 | 0.29 |
ENST00000425853.1
|
KIF9
|
kinesin family member 9 |
| chr18_+_33552667 | 0.27 |
ENST00000333234.5
|
C18orf21
|
chromosome 18 open reading frame 21 |
| chr17_-_34207295 | 0.26 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr15_-_55488817 | 0.23 |
ENST00000569386.1
|
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr5_-_132202329 | 0.23 |
ENST00000378673.2
|
GDF9
|
growth differentiation factor 9 |
| chr14_+_96829886 | 0.22 |
ENST00000556095.1
|
GSKIP
|
GSK3B interacting protein |
| chr1_-_38455650 | 0.20 |
ENST00000448721.2
|
SF3A3
|
splicing factor 3a, subunit 3, 60kDa |
| chr3_-_47324079 | 0.19 |
ENST00000352910.4
|
KIF9
|
kinesin family member 9 |
| chr6_-_33239712 | 0.18 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr12_-_110888103 | 0.16 |
ENST00000426440.1
ENST00000228825.7 |
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr10_+_18948311 | 0.16 |
ENST00000377275.3
|
ARL5B
|
ADP-ribosylation factor-like 5B |
| chr6_-_27440460 | 0.16 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr3_-_47324242 | 0.16 |
ENST00000456548.1
ENST00000432493.1 ENST00000335044.2 ENST00000444589.2 |
KIF9
|
kinesin family member 9 |
| chr22_-_36925124 | 0.16 |
ENST00000457241.1
|
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr8_-_38034192 | 0.15 |
ENST00000520755.1
|
LSM1
|
LSM1 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr19_+_58694742 | 0.15 |
ENST00000597528.1
ENST00000594839.1 |
ZNF274
|
zinc finger protein 274 |
| chr18_-_72265035 | 0.15 |
ENST00000585279.1
ENST00000580048.1 |
LINC00909
|
long intergenic non-protein coding RNA 909 |
| chr3_+_108308513 | 0.15 |
ENST00000361582.3
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr3_+_108308559 | 0.15 |
ENST00000486815.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr6_-_27440837 | 0.14 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chr8_-_38034234 | 0.14 |
ENST00000311351.4
|
LSM1
|
LSM1 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr3_-_28390120 | 0.14 |
ENST00000334100.6
|
AZI2
|
5-azacytidine induced 2 |
| chr15_+_34517251 | 0.13 |
ENST00000559421.1
|
EMC4
|
ER membrane protein complex subunit 4 |
| chr22_-_36924944 | 0.13 |
ENST00000405442.1
ENST00000402116.1 |
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr14_-_68066849 | 0.13 |
ENST00000558493.1
ENST00000561272.1 |
PIGH
|
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr2_+_65454863 | 0.13 |
ENST00000260641.5
|
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast) |
| chr1_-_169337176 | 0.13 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr2_-_201753980 | 0.13 |
ENST00000443398.1
ENST00000286175.8 ENST00000409449.1 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr3_-_101232019 | 0.13 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr11_-_116658695 | 0.13 |
ENST00000429220.1
ENST00000444935.1 |
ZNF259
|
zinc finger protein 259 |
| chr11_-_65625014 | 0.13 |
ENST00000534784.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr1_+_38478432 | 0.13 |
ENST00000537711.1
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr6_-_33239612 | 0.13 |
ENST00000482399.1
ENST00000445902.2 |
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr7_+_108210012 | 0.13 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr17_-_45266542 | 0.13 |
ENST00000531206.1
ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27
|
cell division cycle 27 |
| chr1_+_32687971 | 0.12 |
ENST00000373586.1
|
EIF3I
|
eukaryotic translation initiation factor 3, subunit I |
| chr14_+_35761580 | 0.12 |
ENST00000553809.1
ENST00000555764.1 ENST00000556506.1 |
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr19_+_17326521 | 0.12 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr17_-_7835228 | 0.12 |
ENST00000303731.4
ENST00000571947.1 ENST00000540486.1 ENST00000572656.1 |
TRAPPC1
|
trafficking protein particle complex 1 |
| chr17_+_74723031 | 0.12 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr14_+_96829814 | 0.12 |
ENST00000555181.1
ENST00000553699.1 ENST00000554182.1 |
GSKIP
|
GSK3B interacting protein |
| chr1_+_154947126 | 0.12 |
ENST00000368439.1
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr19_-_47354023 | 0.12 |
ENST00000601649.1
ENST00000599990.1 ENST00000352203.4 |
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr4_+_76439649 | 0.12 |
ENST00000507557.1
|
THAP6
|
THAP domain containing 6 |
| chr2_-_201753859 | 0.11 |
ENST00000409361.1
ENST00000392283.4 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr17_-_65362678 | 0.11 |
ENST00000357146.4
ENST00000356126.3 |
PSMD12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
| chr18_-_72264805 | 0.11 |
ENST00000577806.1
|
LINC00909
|
long intergenic non-protein coding RNA 909 |
| chrX_+_110924346 | 0.11 |
ENST00000371979.3
ENST00000251943.4 ENST00000486353.1 ENST00000394780.3 ENST00000495283.1 |
ALG13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr15_+_74833518 | 0.11 |
ENST00000346246.5
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr5_+_132202252 | 0.11 |
ENST00000378670.3
ENST00000378667.1 ENST00000378665.1 |
UQCRQ
|
ubiquinol-cytochrome c reductase, complex III subunit VII, 9.5kDa |
| chr11_-_9336234 | 0.11 |
ENST00000528080.1
|
TMEM41B
|
transmembrane protein 41B |
| chr22_-_36925186 | 0.11 |
ENST00000541106.1
ENST00000455547.1 ENST00000432675.1 |
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr11_-_116658758 | 0.11 |
ENST00000227322.3
|
ZNF259
|
zinc finger protein 259 |
| chr1_+_169337172 | 0.11 |
ENST00000367807.3
ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr12_+_109490370 | 0.11 |
ENST00000257548.5
ENST00000536723.1 ENST00000536393.1 |
USP30
|
ubiquitin specific peptidase 30 |
| chr2_+_234160340 | 0.11 |
ENST00000417017.1
ENST00000392020.4 ENST00000392018.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr3_-_179322416 | 0.10 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr8_+_146052849 | 0.10 |
ENST00000532777.1
ENST00000325241.6 ENST00000446747.2 ENST00000525266.1 ENST00000544249.1 ENST00000325217.5 ENST00000533314.1 ENST00000527218.1 ENST00000529819.1 ENST00000528372.1 |
ZNF7
|
zinc finger protein 7 |
| chr2_+_217363793 | 0.10 |
ENST00000456586.1
ENST00000598925.1 ENST00000427280.2 |
RPL37A
|
ribosomal protein L37a |
| chr8_-_42698433 | 0.10 |
ENST00000345117.2
ENST00000254250.3 |
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr2_+_9564013 | 0.10 |
ENST00000460593.1
|
CPSF3
|
cleavage and polyadenylation specific factor 3, 73kDa |
| chr14_+_35761540 | 0.10 |
ENST00000261479.4
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr20_+_34287364 | 0.10 |
ENST00000374072.1
ENST00000397416.1 ENST00000336695.4 |
ROMO1
|
reactive oxygen species modulator 1 |
| chr1_-_109618566 | 0.10 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr9_+_131452239 | 0.10 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
| chr2_-_44588694 | 0.10 |
ENST00000409957.1
|
PREPL
|
prolyl endopeptidase-like |
| chr3_+_15468862 | 0.10 |
ENST00000396842.2
|
EAF1
|
ELL associated factor 1 |
| chr4_-_99064387 | 0.10 |
ENST00000295268.3
|
STPG2
|
sperm-tail PG-rich repeat containing 2 |
| chr3_+_119217422 | 0.10 |
ENST00000466984.1
|
TIMMDC1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr12_+_21590549 | 0.10 |
ENST00000545178.1
ENST00000240651.9 |
PYROXD1
|
pyridine nucleotide-disulphide oxidoreductase domain 1 |
| chr17_-_56065540 | 0.10 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr2_+_118846008 | 0.09 |
ENST00000245787.4
|
INSIG2
|
insulin induced gene 2 |
| chr16_-_28222797 | 0.09 |
ENST00000569951.1
ENST00000565698.1 |
XPO6
|
exportin 6 |
| chr15_+_71185148 | 0.09 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr3_-_47324060 | 0.09 |
ENST00000452770.2
|
KIF9
|
kinesin family member 9 |
| chr6_+_28048753 | 0.09 |
ENST00000377325.1
|
ZNF165
|
zinc finger protein 165 |
| chr1_+_38478378 | 0.09 |
ENST00000373014.4
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr2_+_113342011 | 0.09 |
ENST00000324913.5
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr19_-_55690758 | 0.09 |
ENST00000590851.1
|
SYT5
|
synaptotagmin V |
| chr1_+_174969262 | 0.09 |
ENST00000406752.1
ENST00000405362.1 |
CACYBP
|
calcyclin binding protein |
| chr10_-_12237820 | 0.09 |
ENST00000378937.3
ENST00000378927.3 |
NUDT5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr2_-_46844242 | 0.09 |
ENST00000281382.6
|
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr2_-_44588624 | 0.09 |
ENST00000438314.1
ENST00000409936.1 |
PREPL
|
prolyl endopeptidase-like |
| chr4_-_76439483 | 0.09 |
ENST00000380840.2
ENST00000513257.1 ENST00000507014.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr2_-_201753717 | 0.09 |
ENST00000409264.2
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr12_-_51566562 | 0.09 |
ENST00000548108.1
|
TFCP2
|
transcription factor CP2 |
| chr11_-_57102947 | 0.09 |
ENST00000526696.1
|
SSRP1
|
structure specific recognition protein 1 |
| chr2_-_46844159 | 0.09 |
ENST00000474980.1
ENST00000306465.4 |
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr5_+_82373379 | 0.08 |
ENST00000396027.4
ENST00000511817.1 |
XRCC4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr2_-_44588893 | 0.08 |
ENST00000409272.1
ENST00000410081.1 ENST00000541738.1 |
PREPL
|
prolyl endopeptidase-like |
| chr20_+_44486246 | 0.08 |
ENST00000255152.2
ENST00000454862.2 |
ZSWIM3
|
zinc finger, SWIM-type containing 3 |
| chr2_-_44588679 | 0.08 |
ENST00000409411.1
|
PREPL
|
prolyl endopeptidase-like |
| chr11_+_118889456 | 0.08 |
ENST00000528230.1
ENST00000525303.1 ENST00000434101.2 ENST00000359005.4 ENST00000533058.1 |
TRAPPC4
|
trafficking protein particle complex 4 |
| chr10_-_14996017 | 0.08 |
ENST00000378241.1
ENST00000456122.1 ENST00000418843.1 ENST00000378249.1 ENST00000396817.2 ENST00000378255.1 ENST00000378254.1 ENST00000378278.2 ENST00000357717.2 |
DCLRE1C
|
DNA cross-link repair 1C |
| chr11_+_236959 | 0.08 |
ENST00000431206.2
ENST00000528906.1 |
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr12_-_57119068 | 0.08 |
ENST00000546862.1
ENST00000550952.1 ENST00000548563.1 |
NACA
|
nascent polypeptide-associated complex alpha subunit |
| chr19_-_37157713 | 0.08 |
ENST00000592829.1
ENST00000591370.1 ENST00000588268.1 ENST00000360357.4 |
ZNF461
|
zinc finger protein 461 |
| chr17_+_57784826 | 0.08 |
ENST00000262291.4
|
VMP1
|
vacuole membrane protein 1 |
| chr12_+_7053228 | 0.08 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr4_-_76439596 | 0.08 |
ENST00000451788.1
ENST00000512706.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr5_+_82373317 | 0.08 |
ENST00000282268.3
ENST00000338635.6 |
XRCC4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr19_-_51523412 | 0.08 |
ENST00000391805.1
ENST00000599077.1 |
KLK10
|
kallikrein-related peptidase 10 |
| chr22_+_46731676 | 0.08 |
ENST00000424260.2
|
TRMU
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr12_-_57119300 | 0.08 |
ENST00000546917.1
ENST00000454682.1 |
NACA
|
nascent polypeptide-associated complex alpha subunit |
| chr10_-_119806085 | 0.08 |
ENST00000355624.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I) |
| chr15_-_72978490 | 0.08 |
ENST00000311755.3
|
HIGD2B
|
HIG1 hypoxia inducible domain family, member 2B |
| chr15_+_57998923 | 0.08 |
ENST00000380557.4
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr2_-_73964447 | 0.08 |
ENST00000272424.5
ENST00000409716.2 ENST00000318190.7 |
TPRKB
|
TP53RK binding protein |
| chr3_-_10028366 | 0.08 |
ENST00000429759.1
|
EMC3
|
ER membrane protein complex subunit 3 |
| chr16_-_3493528 | 0.08 |
ENST00000301744.4
|
ZNF597
|
zinc finger protein 597 |
| chr6_-_86303523 | 0.08 |
ENST00000513865.1
ENST00000369627.2 ENST00000514419.1 ENST00000509338.1 ENST00000314673.3 ENST00000346348.3 |
SNX14
|
sorting nexin 14 |
| chr3_+_119217376 | 0.08 |
ENST00000494664.1
ENST00000493694.1 |
TIMMDC1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr22_-_30752861 | 0.08 |
ENST00000439242.1
ENST00000215793.8 |
SF3A1
|
splicing factor 3a, subunit 1, 120kDa |
| chr4_-_100815525 | 0.08 |
ENST00000226522.8
ENST00000499666.2 |
LAMTOR3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr20_+_60962143 | 0.08 |
ENST00000343986.4
|
RPS21
|
ribosomal protein S21 |
| chr12_-_58329819 | 0.08 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr5_-_138862326 | 0.08 |
ENST00000330794.4
|
TMEM173
|
transmembrane protein 173 |
| chr2_-_27886676 | 0.08 |
ENST00000337768.5
|
SUPT7L
|
suppressor of Ty 7 (S. cerevisiae)-like |
| chr15_+_40331456 | 0.08 |
ENST00000504245.1
ENST00000560341.1 |
SRP14-AS1
|
SRP14 antisense RNA1 (head to head) |
| chr15_-_65809581 | 0.08 |
ENST00000341861.5
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr12_+_110562135 | 0.08 |
ENST00000361948.4
ENST00000552912.1 ENST00000242591.5 ENST00000546374.1 |
IFT81
|
intraflagellar transport 81 homolog (Chlamydomonas) |
| chr21_-_33984456 | 0.08 |
ENST00000431216.1
ENST00000553001.1 ENST00000440966.1 |
AP000275.65
C21orf59
|
Uncharacterized protein chromosome 21 open reading frame 59 |
| chr2_+_201676256 | 0.07 |
ENST00000452206.1
ENST00000410110.2 ENST00000409600.1 |
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr11_+_118889142 | 0.07 |
ENST00000533632.1
|
TRAPPC4
|
trafficking protein particle complex 4 |
| chr2_-_110371720 | 0.07 |
ENST00000356688.4
|
SEPT10
|
septin 10 |
| chr13_+_50070491 | 0.07 |
ENST00000496612.1
ENST00000357596.3 ENST00000485919.1 ENST00000442195.1 |
PHF11
|
PHD finger protein 11 |
| chr16_-_12009833 | 0.07 |
ENST00000420576.2
|
GSPT1
|
G1 to S phase transition 1 |
| chr2_-_9563469 | 0.07 |
ENST00000484735.1
ENST00000456913.2 |
ITGB1BP1
|
integrin beta 1 binding protein 1 |
| chr1_-_43855444 | 0.07 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr2_+_162016916 | 0.07 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr6_-_153304697 | 0.07 |
ENST00000367241.3
|
FBXO5
|
F-box protein 5 |
| chr13_+_50070077 | 0.07 |
ENST00000378319.3
ENST00000426879.1 |
PHF11
|
PHD finger protein 11 |
| chr14_-_21979428 | 0.07 |
ENST00000538267.1
ENST00000298717.4 |
METTL3
|
methyltransferase like 3 |
| chrX_-_119694538 | 0.07 |
ENST00000371322.5
|
CUL4B
|
cullin 4B |
| chr11_+_71791803 | 0.07 |
ENST00000539271.1
|
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr3_-_28390298 | 0.07 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr3_+_179322573 | 0.07 |
ENST00000493866.1
ENST00000472629.1 ENST00000482604.1 |
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chrX_-_15872914 | 0.07 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr13_+_27998681 | 0.07 |
ENST00000381140.4
|
GTF3A
|
general transcription factor IIIA |
| chr22_+_38349724 | 0.07 |
ENST00000470701.1
|
POLR2F
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr7_-_91509986 | 0.07 |
ENST00000456229.1
ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF
|
mitochondrial transcription termination factor |
| chr2_-_110371777 | 0.07 |
ENST00000397712.2
|
SEPT10
|
septin 10 |
| chr5_+_10250328 | 0.07 |
ENST00000515390.1
|
CCT5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr2_+_46844290 | 0.07 |
ENST00000238892.3
|
CRIPT
|
cysteine-rich PDZ-binding protein |
| chr2_+_230787201 | 0.07 |
ENST00000283946.3
|
FBXO36
|
F-box protein 36 |
| chr18_+_20513278 | 0.07 |
ENST00000327155.5
|
RBBP8
|
retinoblastoma binding protein 8 |
| chr4_+_76439665 | 0.07 |
ENST00000508105.1
ENST00000311638.3 ENST00000380837.3 ENST00000507556.1 ENST00000504190.1 ENST00000507885.1 ENST00000502620.1 ENST00000514480.1 |
THAP6
|
THAP domain containing 6 |
| chr15_-_40331115 | 0.07 |
ENST00000558720.1
|
SRP14
|
signal recognition particle 14kDa (homologous Alu RNA binding protein) |
| chr5_+_76326187 | 0.07 |
ENST00000312916.7
ENST00000506806.1 |
AGGF1
|
angiogenic factor with G patch and FHA domains 1 |
| chr2_+_9563697 | 0.07 |
ENST00000238112.3
|
CPSF3
|
cleavage and polyadenylation specific factor 3, 73kDa |
| chr11_+_47270436 | 0.07 |
ENST00000395397.3
ENST00000405576.1 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr12_-_6798523 | 0.07 |
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384 |
| chr18_+_20513782 | 0.07 |
ENST00000399722.2
ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr1_+_204485503 | 0.07 |
ENST00000367182.3
ENST00000507825.2 |
MDM4
|
Mdm4 p53 binding protein homolog (mouse) |
| chr16_-_2097787 | 0.07 |
ENST00000566380.1
ENST00000219066.1 |
NTHL1
|
nth endonuclease III-like 1 (E. coli) |
| chr10_-_12238071 | 0.07 |
ENST00000491614.1
ENST00000537776.1 |
NUDT5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr22_+_38245414 | 0.07 |
ENST00000381683.6
ENST00000414316.1 ENST00000406934.1 ENST00000451427.1 |
EIF3L
|
eukaryotic translation initiation factor 3, subunit L |
| chr6_+_144164455 | 0.07 |
ENST00000367576.5
|
LTV1
|
LTV1 homolog (S. cerevisiae) |
| chr16_+_19535133 | 0.06 |
ENST00000396212.2
ENST00000381396.5 |
CCP110
|
centriolar coiled coil protein 110kDa |
| chr1_+_24019099 | 0.06 |
ENST00000443624.1
ENST00000458455.1 |
RPL11
|
ribosomal protein L11 |
| chr11_+_237016 | 0.06 |
ENST00000352303.5
|
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr2_-_175351744 | 0.06 |
ENST00000295500.4
ENST00000392552.2 ENST00000392551.2 |
GPR155
|
G protein-coupled receptor 155 |
| chr3_+_28390637 | 0.06 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr3_+_88198838 | 0.06 |
ENST00000318887.3
|
C3orf38
|
chromosome 3 open reading frame 38 |
| chr5_+_79703823 | 0.06 |
ENST00000338008.5
ENST00000510158.1 ENST00000505560.1 |
ZFYVE16
|
zinc finger, FYVE domain containing 16 |
| chr22_+_18111594 | 0.06 |
ENST00000399782.1
|
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr12_-_57119106 | 0.06 |
ENST00000546392.1
|
NACA
|
nascent polypeptide-associated complex alpha subunit |
| chr21_+_44073860 | 0.06 |
ENST00000335512.4
ENST00000539837.1 ENST00000291539.6 ENST00000380328.2 ENST00000398232.3 ENST00000398234.3 ENST00000398236.3 ENST00000328862.6 ENST00000335440.6 ENST00000398225.3 ENST00000398229.3 ENST00000398227.3 |
PDE9A
|
phosphodiesterase 9A |
| chr22_-_18111499 | 0.06 |
ENST00000413576.1
ENST00000399796.2 ENST00000399798.2 ENST00000253413.5 |
ATP6V1E1
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E1 |
| chr21_-_36260980 | 0.06 |
ENST00000344691.4
ENST00000358356.5 |
RUNX1
|
runt-related transcription factor 1 |
| chr5_-_114961858 | 0.06 |
ENST00000282382.4
ENST00000456936.3 ENST00000408996.4 |
TMED7-TICAM2
TMED7
TICAM2
|
TMED7-TICAM2 readthrough transmembrane emp24 protein transport domain containing 7 toll-like receptor adaptor molecule 2 |
| chr15_-_40331356 | 0.06 |
ENST00000560773.1
ENST00000267884.6 |
SRP14
|
signal recognition particle 14kDa (homologous Alu RNA binding protein) |
| chr14_-_69864993 | 0.06 |
ENST00000555373.1
|
ERH
|
enhancer of rudimentary homolog (Drosophila) |
| chr13_+_46039037 | 0.06 |
ENST00000349995.5
|
COG3
|
component of oligomeric golgi complex 3 |
| chr8_-_42698292 | 0.06 |
ENST00000529779.1
|
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr8_+_95565947 | 0.06 |
ENST00000523011.1
|
RP11-267M23.4
|
RP11-267M23.4 |
| chr12_+_56401268 | 0.06 |
ENST00000262032.5
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr12_-_6602955 | 0.06 |
ENST00000543703.1
|
MRPL51
|
mitochondrial ribosomal protein L51 |
| chr7_-_108209897 | 0.06 |
ENST00000313516.5
|
THAP5
|
THAP domain containing 5 |
| chr1_+_168148273 | 0.06 |
ENST00000367830.3
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr15_+_66161792 | 0.06 |
ENST00000564910.1
ENST00000261890.2 |
RAB11A
|
RAB11A, member RAS oncogene family |
| chr17_+_62503147 | 0.06 |
ENST00000553412.1
|
CEP95
|
centrosomal protein 95kDa |
| chr16_+_23652773 | 0.06 |
ENST00000563998.1
ENST00000568589.1 ENST00000568272.1 |
DCTN5
|
dynactin 5 (p25) |
| chr11_-_118122996 | 0.06 |
ENST00000525386.1
ENST00000527472.1 ENST00000278949.4 |
MPZL3
|
myelin protein zero-like 3 |
| chr3_-_108308241 | 0.06 |
ENST00000295746.8
|
KIAA1524
|
KIAA1524 |
| chr18_+_33552597 | 0.06 |
ENST00000269194.6
ENST00000587873.1 |
C18orf21
|
chromosome 18 open reading frame 21 |
| chr5_+_158690089 | 0.06 |
ENST00000296786.6
|
UBLCP1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr20_+_30327063 | 0.06 |
ENST00000300403.6
ENST00000340513.4 |
TPX2
|
TPX2, microtubule-associated |
| chr3_-_180397256 | 0.06 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr17_-_56065484 | 0.06 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr4_+_76439095 | 0.06 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr12_+_7053172 | 0.06 |
ENST00000229281.5
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr15_+_57998821 | 0.06 |
ENST00000299638.3
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr5_-_82373260 | 0.06 |
ENST00000502346.1
|
TMEM167A
|
transmembrane protein 167A |
| chr16_-_12009735 | 0.06 |
ENST00000439887.2
ENST00000434724.2 |
GSPT1
|
G1 to S phase transition 1 |
| chr12_-_57119040 | 0.06 |
ENST00000552540.1
ENST00000393891.4 |
NACA
|
nascent polypeptide-associated complex alpha subunit |
| chr16_+_20817746 | 0.06 |
ENST00000568894.1
|
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr10_-_96122682 | 0.06 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr16_-_47495170 | 0.05 |
ENST00000320640.6
ENST00000544001.2 |
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 0.4 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.2 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.0 | 0.1 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.2 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.0 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.1 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.0 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:2000782 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.7 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.1 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.0 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.0 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.0 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.0 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.0 | GO:2000395 | positive regulation of viral budding via host ESCRT complex(GO:1903774) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.0 | GO:2000426 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.0 | GO:2000619 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.6 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.3 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.0 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.2 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.2 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.0 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.0 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |