Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
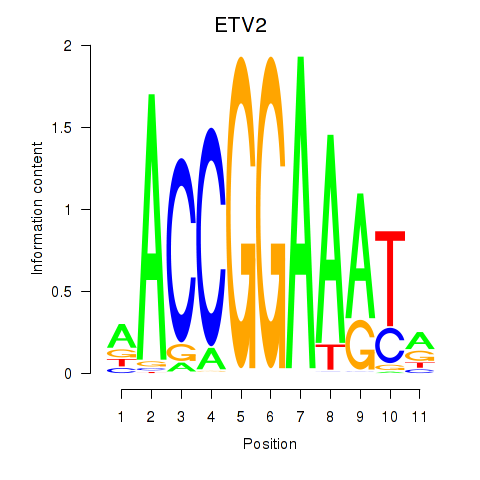
Results for ETV2
Z-value: 0.24
Transcription factors associated with ETV2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV2
|
ENSG00000105672.10 | ETS variant transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV2 | hg19_v2_chr19_+_36132631_36132695 | -0.65 | 3.5e-01 | Click! |
Activity profile of ETV2 motif
Sorted Z-values of ETV2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_71184724 | 0.35 |
ENST00000560604.1
|
THAP10
|
THAP domain containing 10 |
| chr7_+_99006550 | 0.18 |
ENST00000222969.5
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr7_+_99006232 | 0.17 |
ENST00000403633.2
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr12_-_110888103 | 0.17 |
ENST00000426440.1
ENST00000228825.7 |
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr5_+_137801160 | 0.15 |
ENST00000239938.4
|
EGR1
|
early growth response 1 |
| chr1_+_112016414 | 0.14 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr2_+_113342011 | 0.11 |
ENST00000324913.5
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr1_+_38478432 | 0.11 |
ENST00000537711.1
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr15_+_34517251 | 0.10 |
ENST00000559421.1
|
EMC4
|
ER membrane protein complex subunit 4 |
| chr1_+_32687971 | 0.10 |
ENST00000373586.1
|
EIF3I
|
eukaryotic translation initiation factor 3, subunit I |
| chr16_-_3493528 | 0.09 |
ENST00000301744.4
|
ZNF597
|
zinc finger protein 597 |
| chr12_-_123459105 | 0.09 |
ENST00000543935.1
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr15_+_74833518 | 0.09 |
ENST00000346246.5
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr2_-_68290106 | 0.09 |
ENST00000407324.1
ENST00000355848.3 ENST00000409302.1 ENST00000410067.3 |
C1D
|
C1D nuclear receptor corepressor |
| chr15_+_82555169 | 0.09 |
ENST00000565432.1
ENST00000427381.2 |
FAM154B
|
family with sequence similarity 154, member B |
| chr19_-_10305302 | 0.09 |
ENST00000592054.1
|
DNMT1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr1_-_43638168 | 0.09 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr12_-_105629852 | 0.09 |
ENST00000551662.1
ENST00000553097.1 |
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr15_+_71185148 | 0.09 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr6_+_144164455 | 0.09 |
ENST00000367576.5
|
LTV1
|
LTV1 homolog (S. cerevisiae) |
| chr9_+_114393634 | 0.09 |
ENST00000556107.1
ENST00000374294.3 |
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr14_+_32414059 | 0.09 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr3_-_47324008 | 0.08 |
ENST00000425853.1
|
KIF9
|
kinesin family member 9 |
| chr12_-_112847354 | 0.08 |
ENST00000550566.2
ENST00000553213.2 ENST00000424576.2 ENST00000202773.9 |
RPL6
|
ribosomal protein L6 |
| chr2_-_122494487 | 0.08 |
ENST00000451734.1
ENST00000285814.4 |
MKI67IP
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr21_+_27107672 | 0.08 |
ENST00000400075.3
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa |
| chr1_+_38478378 | 0.08 |
ENST00000373014.4
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr17_-_34207295 | 0.08 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr19_+_17326521 | 0.07 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr1_+_84944926 | 0.07 |
ENST00000370656.1
ENST00000370654.5 |
RPF1
|
ribosome production factor 1 homolog (S. cerevisiae) |
| chr8_+_146052849 | 0.07 |
ENST00000532777.1
ENST00000325241.6 ENST00000446747.2 ENST00000525266.1 ENST00000544249.1 ENST00000325217.5 ENST00000533314.1 ENST00000527218.1 ENST00000529819.1 ENST00000528372.1 |
ZNF7
|
zinc finger protein 7 |
| chr2_+_234160340 | 0.07 |
ENST00000417017.1
ENST00000392020.4 ENST00000392018.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr6_-_27440460 | 0.07 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr3_-_28390120 | 0.07 |
ENST00000334100.6
|
AZI2
|
5-azacytidine induced 2 |
| chr3_-_101232019 | 0.07 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr15_+_71184931 | 0.07 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr10_+_18948311 | 0.07 |
ENST00000377275.3
|
ARL5B
|
ADP-ribosylation factor-like 5B |
| chr7_+_139025105 | 0.07 |
ENST00000541170.3
|
C7orf55-LUC7L2
|
C7orf55-LUC7L2 readthrough |
| chr4_-_76912070 | 0.07 |
ENST00000395711.4
ENST00000356260.5 |
SDAD1
|
SDA1 domain containing 1 |
| chr16_-_12009833 | 0.07 |
ENST00000420576.2
|
GSPT1
|
G1 to S phase transition 1 |
| chr14_+_58862630 | 0.07 |
ENST00000360945.2
|
TOMM20L
|
translocase of outer mitochondrial membrane 20 homolog (yeast)-like |
| chr2_-_201753980 | 0.07 |
ENST00000443398.1
ENST00000286175.8 ENST00000409449.1 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr1_-_235324772 | 0.07 |
ENST00000408888.3
|
RBM34
|
RNA binding motif protein 34 |
| chr6_+_31633902 | 0.07 |
ENST00000375865.2
ENST00000375866.2 |
CSNK2B
|
casein kinase 2, beta polypeptide |
| chr3_-_47324242 | 0.07 |
ENST00000456548.1
ENST00000432493.1 ENST00000335044.2 ENST00000444589.2 |
KIF9
|
kinesin family member 9 |
| chr1_-_43637915 | 0.07 |
ENST00000236051.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chrX_+_100646190 | 0.07 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chr14_+_45366518 | 0.06 |
ENST00000557112.1
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr11_+_32605350 | 0.06 |
ENST00000531120.1
ENST00000524896.1 ENST00000323213.5 |
EIF3M
|
eukaryotic translation initiation factor 3, subunit M |
| chr1_+_154947126 | 0.06 |
ENST00000368439.1
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr6_-_27440837 | 0.06 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chr2_+_109065634 | 0.06 |
ENST00000409821.1
|
GCC2
|
GRIP and coiled-coil domain containing 2 |
| chr12_-_51566562 | 0.06 |
ENST00000548108.1
|
TFCP2
|
transcription factor CP2 |
| chr15_-_65809581 | 0.06 |
ENST00000341861.5
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr3_+_23847432 | 0.06 |
ENST00000346855.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr20_+_60962143 | 0.06 |
ENST00000343986.4
|
RPS21
|
ribosomal protein S21 |
| chr12_-_110511424 | 0.06 |
ENST00000548191.1
|
C12orf76
|
chromosome 12 open reading frame 76 |
| chr2_-_201753717 | 0.06 |
ENST00000409264.2
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr4_+_39640787 | 0.06 |
ENST00000532680.1
|
RP11-539G18.2
|
RP11-539G18.2 |
| chr8_+_74903580 | 0.06 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr17_-_65362678 | 0.06 |
ENST00000357146.4
ENST00000356126.3 |
PSMD12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
| chrX_+_110924346 | 0.06 |
ENST00000371979.3
ENST00000251943.4 ENST00000486353.1 ENST00000394780.3 ENST00000495283.1 |
ALG13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr20_-_49575081 | 0.06 |
ENST00000371588.5
ENST00000371582.4 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr3_-_47324079 | 0.06 |
ENST00000352910.4
|
KIF9
|
kinesin family member 9 |
| chr12_+_64846129 | 0.06 |
ENST00000540417.1
ENST00000539810.1 |
TBK1
|
TANK-binding kinase 1 |
| chr2_-_201753859 | 0.06 |
ENST00000409361.1
ENST00000392283.4 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr4_-_39033963 | 0.06 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr17_-_79817091 | 0.06 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr15_+_50716645 | 0.06 |
ENST00000560982.1
|
USP8
|
ubiquitin specific peptidase 8 |
| chr4_-_153700864 | 0.06 |
ENST00000304337.2
|
TIGD4
|
tigger transposable element derived 4 |
| chr12_-_30907822 | 0.06 |
ENST00000540436.1
|
CAPRIN2
|
caprin family member 2 |
| chr4_+_190861941 | 0.06 |
ENST00000226798.4
|
FRG1
|
FSHD region gene 1 |
| chr2_-_133104839 | 0.06 |
ENST00000608279.1
|
RP11-725P16.2
|
RP11-725P16.2 |
| chr4_+_76439649 | 0.06 |
ENST00000507557.1
|
THAP6
|
THAP domain containing 6 |
| chr6_-_108582168 | 0.06 |
ENST00000426155.2
|
SNX3
|
sorting nexin 3 |
| chr7_+_108210012 | 0.06 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr10_+_44101850 | 0.06 |
ENST00000361807.3
ENST00000374437.2 ENST00000430885.1 ENST00000374435.3 |
ZNF485
|
zinc finger protein 485 |
| chr12_+_22852791 | 0.05 |
ENST00000413794.2
|
RP11-114G22.1
|
RP11-114G22.1 |
| chr3_-_69062764 | 0.05 |
ENST00000295571.5
|
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr2_+_65454863 | 0.05 |
ENST00000260641.5
|
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast) |
| chr17_-_8286484 | 0.05 |
ENST00000582556.1
ENST00000584164.1 ENST00000293842.5 ENST00000584343.1 ENST00000578812.1 ENST00000583011.1 |
RPL26
|
ribosomal protein L26 |
| chr6_+_49431073 | 0.05 |
ENST00000335783.3
|
CENPQ
|
centromere protein Q |
| chr16_+_89724188 | 0.05 |
ENST00000301031.4
ENST00000566204.1 ENST00000579310.1 |
SPATA33
|
spermatogenesis associated 33 |
| chr12_-_105630016 | 0.05 |
ENST00000258530.3
|
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr4_+_84377198 | 0.05 |
ENST00000507349.1
ENST00000509970.1 ENST00000505719.1 |
MRPS18C
|
mitochondrial ribosomal protein S18C |
| chr22_-_36925124 | 0.05 |
ENST00000457241.1
|
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr1_-_235324530 | 0.05 |
ENST00000447801.1
ENST00000366606.3 ENST00000429912.1 |
RBM34
|
RNA binding motif protein 34 |
| chr3_+_119217376 | 0.05 |
ENST00000494664.1
ENST00000493694.1 |
TIMMDC1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr6_-_70506963 | 0.05 |
ENST00000370577.3
|
LMBRD1
|
LMBR1 domain containing 1 |
| chr6_-_33239712 | 0.05 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr9_+_114393581 | 0.05 |
ENST00000313525.3
|
DNAJC25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr12_+_120875910 | 0.05 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chrX_+_17755563 | 0.05 |
ENST00000380045.3
ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr1_-_38455650 | 0.05 |
ENST00000448721.2
|
SF3A3
|
splicing factor 3a, subunit 3, 60kDa |
| chr8_-_125551278 | 0.05 |
ENST00000519232.1
ENST00000523888.1 ENST00000522810.1 ENST00000519548.1 ENST00000517678.1 ENST00000605953.1 ENST00000276692.6 |
TATDN1
|
TatD DNase domain containing 1 |
| chr12_-_13248598 | 0.05 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr19_-_37157713 | 0.05 |
ENST00000592829.1
ENST00000591370.1 ENST00000588268.1 ENST00000360357.4 |
ZNF461
|
zinc finger protein 461 |
| chr6_+_31633833 | 0.05 |
ENST00000375882.2
ENST00000375880.2 |
CSNK2B
CSNK2B-LY6G5B-1181
|
casein kinase 2, beta polypeptide Uncharacterized protein |
| chr2_+_118846008 | 0.05 |
ENST00000245787.4
|
INSIG2
|
insulin induced gene 2 |
| chr9_+_33264861 | 0.05 |
ENST00000223500.8
|
CHMP5
|
charged multivesicular body protein 5 |
| chr21_+_44073860 | 0.05 |
ENST00000335512.4
ENST00000539837.1 ENST00000291539.6 ENST00000380328.2 ENST00000398232.3 ENST00000398234.3 ENST00000398236.3 ENST00000328862.6 ENST00000335440.6 ENST00000398225.3 ENST00000398229.3 ENST00000398227.3 |
PDE9A
|
phosphodiesterase 9A |
| chr2_-_110371720 | 0.05 |
ENST00000356688.4
|
SEPT10
|
septin 10 |
| chr5_-_132202329 | 0.05 |
ENST00000378673.2
|
GDF9
|
growth differentiation factor 9 |
| chr19_+_17326141 | 0.05 |
ENST00000445667.2
ENST00000263897.5 |
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr20_-_49575058 | 0.05 |
ENST00000371584.4
ENST00000371583.5 ENST00000413082.1 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr2_-_44588624 | 0.05 |
ENST00000438314.1
ENST00000409936.1 |
PREPL
|
prolyl endopeptidase-like |
| chr8_+_182368 | 0.05 |
ENST00000522866.1
ENST00000398612.1 |
ZNF596
|
zinc finger protein 596 |
| chr1_+_24018269 | 0.05 |
ENST00000374550.3
|
RPL11
|
ribosomal protein L11 |
| chr1_-_55266865 | 0.05 |
ENST00000371274.4
|
TTC22
|
tetratricopeptide repeat domain 22 |
| chr5_+_158690089 | 0.05 |
ENST00000296786.6
|
UBLCP1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr12_-_30907862 | 0.05 |
ENST00000541765.1
ENST00000537108.1 |
CAPRIN2
|
caprin family member 2 |
| chr8_-_101734170 | 0.05 |
ENST00000522387.1
ENST00000518196.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr9_+_131452239 | 0.05 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
| chr2_-_110371412 | 0.05 |
ENST00000415095.1
ENST00000334001.6 ENST00000437928.1 ENST00000493445.1 ENST00000397714.2 ENST00000461295.1 |
SEPT10
|
septin 10 |
| chr12_+_109490370 | 0.05 |
ENST00000257548.5
ENST00000536723.1 ENST00000536393.1 |
USP30
|
ubiquitin specific peptidase 30 |
| chr15_+_80364901 | 0.05 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr14_-_71609965 | 0.05 |
ENST00000602957.1
|
RP6-91H8.3
|
RP6-91H8.3 |
| chr1_-_17380630 | 0.05 |
ENST00000375499.3
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr11_+_18344106 | 0.05 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr2_+_85839218 | 0.05 |
ENST00000448971.1
ENST00000442708.1 ENST00000450066.2 |
USP39
|
ubiquitin specific peptidase 39 |
| chr9_+_33265011 | 0.05 |
ENST00000419016.2
|
CHMP5
|
charged multivesicular body protein 5 |
| chr12_-_30907749 | 0.05 |
ENST00000542550.1
ENST00000540584.1 |
CAPRIN2
|
caprin family member 2 |
| chr10_-_18948208 | 0.05 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr19_-_8942962 | 0.05 |
ENST00000601372.1
|
ZNF558
|
zinc finger protein 558 |
| chr3_+_14219858 | 0.05 |
ENST00000306024.3
|
LSM3
|
LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr1_-_63988846 | 0.05 |
ENST00000283568.8
ENST00000371092.3 ENST00000271002.10 |
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr16_-_12009735 | 0.05 |
ENST00000439887.2
ENST00000434724.2 |
GSPT1
|
G1 to S phase transition 1 |
| chr3_-_28390298 | 0.05 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr1_-_109618566 | 0.05 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr5_+_82373379 | 0.04 |
ENST00000396027.4
ENST00000511817.1 |
XRCC4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr4_-_492891 | 0.04 |
ENST00000338977.5
ENST00000511833.2 |
ZNF721
|
zinc finger protein 721 |
| chr11_+_18343800 | 0.04 |
ENST00000453096.2
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr17_-_41132088 | 0.04 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr3_-_180397256 | 0.04 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr8_-_55014336 | 0.04 |
ENST00000343231.6
|
LYPLA1
|
lysophospholipase I |
| chrX_-_131352152 | 0.04 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr1_-_169337176 | 0.04 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr16_+_20912075 | 0.04 |
ENST00000219168.4
|
LYRM1
|
LYR motif containing 1 |
| chr2_-_110371777 | 0.04 |
ENST00000397712.2
|
SEPT10
|
septin 10 |
| chr12_-_6602955 | 0.04 |
ENST00000543703.1
|
MRPL51
|
mitochondrial ribosomal protein L51 |
| chr14_+_35452104 | 0.04 |
ENST00000216774.6
ENST00000546080.1 |
SRP54
|
signal recognition particle 54kDa |
| chr12_+_7053228 | 0.04 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr5_+_82373317 | 0.04 |
ENST00000282268.3
ENST00000338635.6 |
XRCC4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr10_-_98347063 | 0.04 |
ENST00000443638.1
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr10_+_103912137 | 0.04 |
ENST00000603742.1
ENST00000488254.2 ENST00000461421.1 ENST00000476468.1 ENST00000370007.5 |
NOLC1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr12_-_6798410 | 0.04 |
ENST00000361959.3
ENST00000436774.2 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chr11_+_9482551 | 0.04 |
ENST00000438144.2
ENST00000526657.1 ENST00000299606.2 ENST00000534265.1 ENST00000412390.2 |
ZNF143
|
zinc finger protein 143 |
| chr10_-_22292613 | 0.04 |
ENST00000376980.3
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr11_+_71791803 | 0.04 |
ENST00000539271.1
|
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr7_-_108209897 | 0.04 |
ENST00000313516.5
|
THAP5
|
THAP domain containing 5 |
| chrX_-_119695279 | 0.04 |
ENST00000336592.6
|
CUL4B
|
cullin 4B |
| chr4_+_76439095 | 0.04 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr7_-_108210048 | 0.04 |
ENST00000415914.3
ENST00000438865.1 |
THAP5
|
THAP domain containing 5 |
| chr6_-_153323801 | 0.04 |
ENST00000367233.5
ENST00000367231.5 ENST00000367230.1 |
MTRF1L
|
mitochondrial translational release factor 1-like |
| chr5_+_179135246 | 0.04 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr7_+_30589829 | 0.04 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr14_-_21979428 | 0.04 |
ENST00000538267.1
ENST00000298717.4 |
METTL3
|
methyltransferase like 3 |
| chr16_-_28222797 | 0.04 |
ENST00000569951.1
ENST00000565698.1 |
XPO6
|
exportin 6 |
| chrX_+_70586082 | 0.04 |
ENST00000373790.4
ENST00000449580.1 ENST00000423759.1 |
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr2_+_162016827 | 0.04 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr7_-_91509986 | 0.04 |
ENST00000456229.1
ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF
|
mitochondrial transcription termination factor |
| chr1_+_40505891 | 0.04 |
ENST00000372797.3
ENST00000372802.1 ENST00000449311.1 |
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr3_+_139062838 | 0.04 |
ENST00000310776.4
ENST00000465056.1 ENST00000465373.1 |
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr17_-_56065540 | 0.04 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr2_-_44588694 | 0.04 |
ENST00000409957.1
|
PREPL
|
prolyl endopeptidase-like |
| chr19_-_6279932 | 0.04 |
ENST00000252674.7
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
| chr12_-_6798523 | 0.04 |
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384 |
| chr19_+_51774540 | 0.04 |
ENST00000600813.1
|
CTD-3187F8.11
|
CTD-3187F8.11 |
| chr7_-_151574191 | 0.04 |
ENST00000287878.4
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr8_+_182422 | 0.04 |
ENST00000518414.1
ENST00000521270.1 ENST00000518320.2 |
ZNF596
|
zinc finger protein 596 |
| chr5_+_76326187 | 0.04 |
ENST00000312916.7
ENST00000506806.1 |
AGGF1
|
angiogenic factor with G patch and FHA domains 1 |
| chr2_-_44588893 | 0.04 |
ENST00000409272.1
ENST00000410081.1 ENST00000541738.1 |
PREPL
|
prolyl endopeptidase-like |
| chr1_-_13840483 | 0.04 |
ENST00000376085.3
|
LRRC38
|
leucine rich repeat containing 38 |
| chr3_+_32726774 | 0.04 |
ENST00000538368.1
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr3_+_32726620 | 0.04 |
ENST00000331889.6
ENST00000328834.5 |
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr3_+_197464046 | 0.04 |
ENST00000428738.1
|
FYTTD1
|
forty-two-three domain containing 1 |
| chr4_-_76439596 | 0.04 |
ENST00000451788.1
ENST00000512706.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr2_-_63815628 | 0.04 |
ENST00000409562.3
|
WDPCP
|
WD repeat containing planar cell polarity effector |
| chr13_+_44453688 | 0.04 |
ENST00000425906.1
|
LACC1
|
laccase (multicopper oxidoreductase) domain containing 1 |
| chr15_-_55488817 | 0.04 |
ENST00000569386.1
|
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr5_+_96079240 | 0.04 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr2_+_38152462 | 0.04 |
ENST00000354545.2
|
RMDN2
|
regulator of microtubule dynamics 2 |
| chr10_-_127408011 | 0.04 |
ENST00000531977.1
ENST00000527483.1 ENST00000525909.1 ENST00000528844.1 ENST00000423178.2 |
RP11-383C5.4
|
RP11-383C5.4 |
| chr18_+_20513278 | 0.04 |
ENST00000327155.5
|
RBBP8
|
retinoblastoma binding protein 8 |
| chr15_+_69706643 | 0.04 |
ENST00000352331.4
ENST00000260363.4 |
KIF23
|
kinesin family member 23 |
| chr6_-_108395907 | 0.04 |
ENST00000193322.3
|
OSTM1
|
osteopetrosis associated transmembrane protein 1 |
| chr3_+_100428188 | 0.04 |
ENST00000418917.2
ENST00000490574.1 |
TFG
|
TRK-fused gene |
| chr18_+_20513782 | 0.04 |
ENST00000399722.2
ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr16_+_20817746 | 0.04 |
ENST00000568894.1
|
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr2_+_162016916 | 0.04 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr5_+_147763539 | 0.04 |
ENST00000296701.6
ENST00000394370.3 |
FBXO38
|
F-box protein 38 |
| chr11_+_113185778 | 0.04 |
ENST00000524580.2
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr10_+_7830125 | 0.04 |
ENST00000335698.4
ENST00000541227.1 |
ATP5C1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr11_-_59436453 | 0.04 |
ENST00000300146.9
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr17_-_56065484 | 0.04 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr3_-_37218023 | 0.04 |
ENST00000416425.1
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr12_-_57119068 | 0.04 |
ENST00000546862.1
ENST00000550952.1 ENST00000548563.1 |
NACA
|
nascent polypeptide-associated complex alpha subunit |
| chr12_-_64784306 | 0.04 |
ENST00000543259.1
|
C12orf56
|
chromosome 12 open reading frame 56 |
| chr1_-_43855444 | 0.04 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr1_-_231376836 | 0.04 |
ENST00000451322.1
|
C1orf131
|
chromosome 1 open reading frame 131 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0045360 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.1 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.0 | 0.1 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.1 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.0 | 0.1 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.0 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.0 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.0 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.0 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.0 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.0 | GO:0009452 | 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 | 0.0 | GO:0021764 | amygdala development(GO:0021764) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.1 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.0 | GO:0034708 | methyltransferase complex(GO:0034708) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.0 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.0 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.0 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.0 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.0 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.0 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.0 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.0 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |