Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
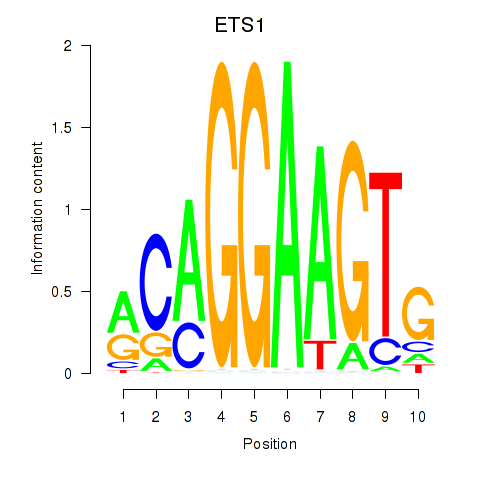
Results for ETS1
Z-value: 0.44
Transcription factors associated with ETS1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETS1
|
ENSG00000134954.10 | ETS proto-oncogene 1, transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETS1 | hg19_v2_chr11_-_128392085_128392232 | 0.35 | 6.5e-01 | Click! |
Activity profile of ETS1 motif
Sorted Z-values of ETS1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_144665237 | 0.20 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr19_-_10305302 | 0.20 |
ENST00000592054.1
|
DNMT1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr9_-_115249484 | 0.13 |
ENST00000457681.1
|
C9orf147
|
chromosome 9 open reading frame 147 |
| chr16_-_31085033 | 0.13 |
ENST00000414399.1
|
ZNF668
|
zinc finger protein 668 |
| chr6_-_134861089 | 0.13 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr5_-_126409159 | 0.13 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr1_+_84767289 | 0.12 |
ENST00000394834.3
ENST00000370669.1 |
SAMD13
|
sterile alpha motif domain containing 13 |
| chr2_-_191399073 | 0.11 |
ENST00000421038.1
|
TMEM194B
|
transmembrane protein 194B |
| chr15_+_23810903 | 0.11 |
ENST00000564592.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr10_-_18948208 | 0.11 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr1_-_151254362 | 0.11 |
ENST00000447795.2
|
RP11-126K1.2
|
Uncharacterized protein |
| chr8_-_100025238 | 0.11 |
ENST00000521696.1
|
RP11-410L14.2
|
RP11-410L14.2 |
| chr16_-_30394143 | 0.11 |
ENST00000321367.3
ENST00000571393.1 |
SEPT1
|
septin 1 |
| chr22_+_38004942 | 0.10 |
ENST00000439161.1
ENST00000449944.1 ENST00000411501.1 ENST00000453208.1 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr11_-_118889323 | 0.10 |
ENST00000527673.1
|
RPS25
|
ribosomal protein S25 |
| chr17_+_38673270 | 0.10 |
ENST00000578280.1
|
RP5-1028K7.2
|
RP5-1028K7.2 |
| chr10_-_127408011 | 0.10 |
ENST00000531977.1
ENST00000527483.1 ENST00000525909.1 ENST00000528844.1 ENST00000423178.2 |
RP11-383C5.4
|
RP11-383C5.4 |
| chr1_+_112016414 | 0.10 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr16_+_20817761 | 0.09 |
ENST00000568046.1
ENST00000261377.6 |
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr14_+_50159813 | 0.09 |
ENST00000359332.2
ENST00000553274.1 ENST00000557128.1 |
KLHDC1
|
kelch domain containing 1 |
| chr12_-_57030096 | 0.09 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr2_+_136499287 | 0.09 |
ENST00000415164.1
|
UBXN4
|
UBX domain protein 4 |
| chr2_-_230786378 | 0.09 |
ENST00000430954.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr22_+_27068704 | 0.09 |
ENST00000444388.1
ENST00000450963.1 ENST00000449017.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr4_-_140005443 | 0.08 |
ENST00000510408.1
ENST00000420916.2 ENST00000358635.3 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr4_-_156297949 | 0.08 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr16_+_20817953 | 0.08 |
ENST00000568647.1
|
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr7_-_130597935 | 0.08 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr16_-_28223229 | 0.08 |
ENST00000566073.1
|
XPO6
|
exportin 6 |
| chr19_-_51289436 | 0.08 |
ENST00000562076.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr15_+_23810853 | 0.08 |
ENST00000568252.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr14_+_32414059 | 0.08 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr1_-_52520828 | 0.08 |
ENST00000610127.1
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr12_-_93835665 | 0.08 |
ENST00000552442.1
ENST00000550657.1 |
UBE2N
|
ubiquitin-conjugating enzyme E2N |
| chr15_+_44092784 | 0.08 |
ENST00000458412.1
|
HYPK
|
huntingtin interacting protein K |
| chr15_+_90895471 | 0.08 |
ENST00000354377.3
ENST00000379090.5 |
ZNF774
|
zinc finger protein 774 |
| chr14_+_102276192 | 0.08 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr1_+_169337412 | 0.08 |
ENST00000426663.1
|
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr11_+_113185292 | 0.07 |
ENST00000429951.1
ENST00000442859.1 ENST00000531164.1 ENST00000529850.1 ENST00000314756.3 ENST00000525965.1 |
TTC12
|
tetratricopeptide repeat domain 12 |
| chr10_-_43633747 | 0.07 |
ENST00000609407.1
|
RP11-351D16.3
|
RP11-351D16.3 |
| chr1_-_1590418 | 0.07 |
ENST00000341028.7
|
CDK11B
|
cyclin-dependent kinase 11B |
| chr17_+_74261413 | 0.07 |
ENST00000587913.1
|
UBALD2
|
UBA-like domain containing 2 |
| chr2_-_32390801 | 0.07 |
ENST00000608489.1
|
RP11-563N4.1
|
RP11-563N4.1 |
| chr16_+_20817746 | 0.07 |
ENST00000568894.1
|
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr15_+_83209620 | 0.07 |
ENST00000568285.1
|
RP11-379H8.1
|
Uncharacterized protein |
| chr19_-_2085323 | 0.07 |
ENST00000591638.1
|
MOB3A
|
MOB kinase activator 3A |
| chr15_-_65809625 | 0.07 |
ENST00000560436.1
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr12_-_80084333 | 0.07 |
ENST00000552637.1
|
PAWR
|
PRKC, apoptosis, WT1, regulator |
| chr1_-_25558984 | 0.07 |
ENST00000236273.4
|
SYF2
|
SYF2 pre-mRNA-splicing factor |
| chr22_+_38004832 | 0.07 |
ENST00000405147.3
ENST00000429218.1 ENST00000325180.8 ENST00000337437.4 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr2_+_138721850 | 0.07 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr9_-_26892765 | 0.07 |
ENST00000520187.1
ENST00000333916.5 |
CAAP1
|
caspase activity and apoptosis inhibitor 1 |
| chr4_-_83812157 | 0.07 |
ENST00000513323.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr16_-_3149278 | 0.07 |
ENST00000575108.1
ENST00000576483.1 ENST00000538082.2 ENST00000576985.1 |
ZSCAN10
|
zinc finger and SCAN domain containing 10 |
| chr16_-_3493528 | 0.07 |
ENST00000301744.4
|
ZNF597
|
zinc finger protein 597 |
| chr15_-_90233907 | 0.07 |
ENST00000561224.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr19_+_39936207 | 0.07 |
ENST00000594729.1
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr6_-_41040049 | 0.07 |
ENST00000471367.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr11_-_62389577 | 0.07 |
ENST00000534715.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr14_+_77843459 | 0.07 |
ENST00000216471.4
|
SAMD15
|
sterile alpha motif domain containing 15 |
| chr21_-_27107283 | 0.07 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr14_-_24898731 | 0.07 |
ENST00000267406.6
|
CBLN3
|
cerebellin 3 precursor |
| chr16_-_30393752 | 0.07 |
ENST00000566517.1
ENST00000605106.1 |
SEPT1
SEPT1
|
septin 1 Uncharacterized protein |
| chr1_+_246729815 | 0.07 |
ENST00000366511.1
|
CNST
|
consortin, connexin sorting protein |
| chr14_-_69658127 | 0.06 |
ENST00000556182.1
|
RP11-363J20.1
|
RP11-363J20.1 |
| chr17_-_62503015 | 0.06 |
ENST00000581806.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr6_+_33257346 | 0.06 |
ENST00000374606.5
ENST00000374610.2 ENST00000374607.1 |
PFDN6
|
prefoldin subunit 6 |
| chr11_+_64008443 | 0.06 |
ENST00000309366.4
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr19_+_36393367 | 0.06 |
ENST00000246551.4
|
HCST
|
hematopoietic cell signal transducer |
| chr9_-_134145880 | 0.06 |
ENST00000372269.3
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78, member A |
| chr14_+_58862630 | 0.06 |
ENST00000360945.2
|
TOMM20L
|
translocase of outer mitochondrial membrane 20 homolog (yeast)-like |
| chr4_-_83295296 | 0.06 |
ENST00000507010.1
ENST00000503822.1 |
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr6_-_31620095 | 0.06 |
ENST00000424176.1
ENST00000456622.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr5_-_102455801 | 0.06 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr2_-_99224915 | 0.06 |
ENST00000328709.3
ENST00000409997.1 |
COA5
|
cytochrome c oxidase assembly factor 5 |
| chr13_+_111805980 | 0.06 |
ENST00000491775.1
ENST00000466143.1 ENST00000544132.1 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr17_+_45908974 | 0.06 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr11_+_45168182 | 0.06 |
ENST00000526442.1
|
PRDM11
|
PR domain containing 11 |
| chr2_-_230786032 | 0.06 |
ENST00000428959.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr11_-_9336117 | 0.06 |
ENST00000527813.1
ENST00000533723.1 |
TMEM41B
|
transmembrane protein 41B |
| chr9_-_14322319 | 0.06 |
ENST00000606230.1
|
NFIB
|
nuclear factor I/B |
| chr1_-_161015663 | 0.06 |
ENST00000534633.1
|
USF1
|
upstream transcription factor 1 |
| chr11_-_125773085 | 0.06 |
ENST00000227474.3
ENST00000534158.1 ENST00000529801.1 |
PUS3
|
pseudouridylate synthase 3 |
| chr11_+_64008525 | 0.06 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr4_+_79567057 | 0.06 |
ENST00000503259.1
ENST00000507802.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr3_-_15469045 | 0.06 |
ENST00000450816.2
|
METTL6
|
methyltransferase like 6 |
| chr6_+_134274354 | 0.06 |
ENST00000367869.1
|
TBPL1
|
TBP-like 1 |
| chr12_+_7055631 | 0.06 |
ENST00000543115.1
ENST00000399448.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr3_+_9774164 | 0.06 |
ENST00000426583.1
|
BRPF1
|
bromodomain and PHD finger containing, 1 |
| chr15_+_71184931 | 0.06 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr15_+_90234028 | 0.06 |
ENST00000268130.7
ENST00000560294.1 ENST00000558000.1 |
WDR93
|
WD repeat domain 93 |
| chr5_+_180650271 | 0.05 |
ENST00000351937.5
ENST00000315073.5 |
TRIM41
|
tripartite motif containing 41 |
| chr5_-_87564620 | 0.05 |
ENST00000506536.1
ENST00000512429.1 ENST00000514135.1 ENST00000296595.6 ENST00000509387.1 |
TMEM161B
|
transmembrane protein 161B |
| chr1_+_144339738 | 0.05 |
ENST00000538264.1
|
AL592284.1
|
Protein LOC642441 |
| chr6_+_43968306 | 0.05 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr14_-_69864993 | 0.05 |
ENST00000555373.1
|
ERH
|
enhancer of rudimentary homolog (Drosophila) |
| chr4_+_190861993 | 0.05 |
ENST00000524583.1
ENST00000531991.2 |
FRG1
|
FSHD region gene 1 |
| chr11_-_61197406 | 0.05 |
ENST00000541963.1
ENST00000477890.2 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr5_+_68530697 | 0.05 |
ENST00000256443.3
ENST00000514676.1 |
CDK7
|
cyclin-dependent kinase 7 |
| chr21_-_27107198 | 0.05 |
ENST00000400094.1
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr3_-_57113281 | 0.05 |
ENST00000468466.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr8_-_131028782 | 0.05 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr1_+_70820451 | 0.05 |
ENST00000361764.4
ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3
|
HERV-H LTR-associating 3 |
| chr12_-_123634449 | 0.05 |
ENST00000542210.1
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr6_-_31619697 | 0.05 |
ENST00000434444.1
|
BAG6
|
BCL2-associated athanogene 6 |
| chr11_-_61196858 | 0.05 |
ENST00000413184.2
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr2_-_37193606 | 0.05 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr5_-_118324200 | 0.05 |
ENST00000515439.3
ENST00000510708.1 |
DTWD2
|
DTW domain containing 2 |
| chr15_+_101402041 | 0.05 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr1_+_112939121 | 0.05 |
ENST00000441739.1
|
CTTNBP2NL
|
CTTNBP2 N-terminal like |
| chr12_-_9102224 | 0.05 |
ENST00000543845.1
ENST00000544245.1 |
M6PR
|
mannose-6-phosphate receptor (cation dependent) |
| chr1_+_222886694 | 0.05 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr2_+_219724544 | 0.05 |
ENST00000233948.3
|
WNT6
|
wingless-type MMTV integration site family, member 6 |
| chr3_-_170626376 | 0.05 |
ENST00000487522.1
ENST00000474417.1 |
EIF5A2
|
eukaryotic translation initiation factor 5A2 |
| chr5_+_80597419 | 0.05 |
ENST00000254037.2
ENST00000407610.3 ENST00000380199.5 |
ZCCHC9
|
zinc finger, CCHC domain containing 9 |
| chr3_-_170626418 | 0.05 |
ENST00000474096.1
ENST00000295822.2 |
EIF5A2
|
eukaryotic translation initiation factor 5A2 |
| chr21_-_33984888 | 0.05 |
ENST00000382549.4
ENST00000540881.1 |
C21orf59
|
chromosome 21 open reading frame 59 |
| chr6_-_41040195 | 0.05 |
ENST00000463088.1
ENST00000469104.1 ENST00000486443.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr5_+_102455853 | 0.05 |
ENST00000515845.1
ENST00000321521.9 ENST00000507921.1 |
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr16_+_80574854 | 0.05 |
ENST00000305904.6
ENST00000568035.1 |
DYNLRB2
|
dynein, light chain, roadblock-type 2 |
| chr7_-_91875109 | 0.05 |
ENST00000412043.2
ENST00000430102.1 ENST00000425073.1 ENST00000394503.2 ENST00000454017.1 ENST00000440209.1 ENST00000413688.1 ENST00000452773.1 ENST00000433016.1 ENST00000394505.2 ENST00000422347.1 ENST00000458493.1 ENST00000425919.1 |
KRIT1
|
KRIT1, ankyrin repeat containing |
| chr12_-_6798025 | 0.05 |
ENST00000542351.1
ENST00000538829.1 |
ZNF384
|
zinc finger protein 384 |
| chr5_-_141060389 | 0.05 |
ENST00000504448.1
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr4_-_15683230 | 0.05 |
ENST00000515679.1
|
FBXL5
|
F-box and leucine-rich repeat protein 5 |
| chr2_+_231191875 | 0.05 |
ENST00000444636.1
ENST00000415673.2 ENST00000243810.6 ENST00000396563.4 |
SP140L
|
SP140 nuclear body protein-like |
| chr2_+_162016827 | 0.05 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr6_-_31633624 | 0.05 |
ENST00000375895.2
ENST00000375900.4 |
GPANK1
|
G patch domain and ankyrin repeats 1 |
| chr11_+_113185778 | 0.05 |
ENST00000524580.2
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr2_-_153573887 | 0.05 |
ENST00000493468.2
ENST00000545856.1 |
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr10_-_95462265 | 0.05 |
ENST00000536233.1
ENST00000359204.4 ENST00000371430.2 ENST00000394100.2 |
FRA10AC1
|
fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 |
| chr6_+_33257427 | 0.05 |
ENST00000463584.1
|
PFDN6
|
prefoldin subunit 6 |
| chr21_+_18885318 | 0.05 |
ENST00000400166.1
|
CXADR
|
coxsackie virus and adenovirus receptor |
| chr21_-_33984456 | 0.05 |
ENST00000431216.1
ENST00000553001.1 ENST00000440966.1 |
AP000275.65
C21orf59
|
Uncharacterized protein chromosome 21 open reading frame 59 |
| chr19_-_58514129 | 0.05 |
ENST00000552184.1
ENST00000546715.1 ENST00000536132.1 ENST00000547828.1 ENST00000547121.1 ENST00000551380.1 |
ZNF606
|
zinc finger protein 606 |
| chr7_+_138915102 | 0.05 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr2_+_217363559 | 0.05 |
ENST00000600880.1
ENST00000446558.1 |
RPL37A
|
ribosomal protein L37a |
| chr11_+_117103333 | 0.05 |
ENST00000534428.1
|
RNF214
|
ring finger protein 214 |
| chr1_-_1655713 | 0.05 |
ENST00000401096.2
ENST00000404249.3 ENST00000357760.2 ENST00000358779.5 ENST00000378633.1 ENST00000378635.3 |
CDK11A
|
cyclin-dependent kinase 11A |
| chr12_-_118796971 | 0.05 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr14_+_45366518 | 0.05 |
ENST00000557112.1
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr6_-_138866823 | 0.05 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr17_+_71228843 | 0.05 |
ENST00000582391.1
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chr5_-_138862326 | 0.05 |
ENST00000330794.4
|
TMEM173
|
transmembrane protein 173 |
| chr1_-_113160826 | 0.05 |
ENST00000538187.1
ENST00000369664.1 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr14_+_57735636 | 0.05 |
ENST00000556995.1
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr8_+_72755796 | 0.05 |
ENST00000524152.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr17_-_43510282 | 0.05 |
ENST00000290470.3
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr20_-_49575058 | 0.05 |
ENST00000371584.4
ENST00000371583.5 ENST00000413082.1 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr10_+_1102303 | 0.05 |
ENST00000381329.1
|
WDR37
|
WD repeat domain 37 |
| chr8_+_110552046 | 0.05 |
ENST00000529931.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr19_+_6464502 | 0.05 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr3_+_49977894 | 0.05 |
ENST00000433811.1
|
RBM6
|
RNA binding motif protein 6 |
| chr13_+_28527647 | 0.05 |
ENST00000567234.1
|
LINC00543
|
long intergenic non-protein coding RNA 543 |
| chr14_+_70233810 | 0.05 |
ENST00000394366.2
ENST00000553548.1 ENST00000553369.1 ENST00000557154.1 ENST00000451983.2 ENST00000553635.1 |
SRSF5
|
serine/arginine-rich splicing factor 5 |
| chr1_-_242162375 | 0.04 |
ENST00000357246.3
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma |
| chr9_-_26892342 | 0.04 |
ENST00000535437.1
|
CAAP1
|
caspase activity and apoptosis inhibitor 1 |
| chrX_-_11129229 | 0.04 |
ENST00000608176.1
ENST00000433747.2 ENST00000608576.1 ENST00000608916.1 |
RP11-120D5.1
|
RP11-120D5.1 |
| chr5_+_179220979 | 0.04 |
ENST00000292596.10
ENST00000401985.3 |
LTC4S
|
leukotriene C4 synthase |
| chr5_+_138677515 | 0.04 |
ENST00000265192.4
ENST00000511706.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr6_-_160210692 | 0.04 |
ENST00000538128.1
ENST00000537390.1 |
TCP1
|
t-complex 1 |
| chr5_+_55354822 | 0.04 |
ENST00000511861.1
|
CTD-2227I18.1
|
CTD-2227I18.1 |
| chr4_-_140005341 | 0.04 |
ENST00000379549.2
ENST00000512627.1 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr2_+_170440902 | 0.04 |
ENST00000448752.2
ENST00000418888.1 ENST00000414307.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr8_-_28347737 | 0.04 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr20_-_35807741 | 0.04 |
ENST00000434295.1
ENST00000441008.2 ENST00000400441.3 ENST00000343811.4 |
MROH8
|
maestro heat-like repeat family member 8 |
| chr20_-_49575081 | 0.04 |
ENST00000371588.5
ENST00000371582.4 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr15_-_65810042 | 0.04 |
ENST00000321147.6
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr12_-_42719885 | 0.04 |
ENST00000552673.1
ENST00000266529.3 ENST00000552235.1 |
ZCRB1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr7_-_91875358 | 0.04 |
ENST00000458177.1
ENST00000394507.1 ENST00000340022.2 ENST00000444960.1 |
KRIT1
|
KRIT1, ankyrin repeat containing |
| chr6_+_75994709 | 0.04 |
ENST00000438676.1
ENST00000607221.1 |
RP1-234P15.4
|
RP1-234P15.4 |
| chr5_-_138861926 | 0.04 |
ENST00000510817.1
|
TMEM173
|
transmembrane protein 173 |
| chr13_-_46626847 | 0.04 |
ENST00000242848.4
ENST00000282007.3 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr6_-_41040268 | 0.04 |
ENST00000373154.2
ENST00000244558.9 ENST00000464633.1 ENST00000424266.2 ENST00000479950.1 ENST00000482515.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr16_-_57219721 | 0.04 |
ENST00000562406.1
ENST00000568671.1 ENST00000567044.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr3_+_14693247 | 0.04 |
ENST00000383794.3
ENST00000303688.7 |
CCDC174
|
coiled-coil domain containing 174 |
| chr18_+_77867177 | 0.04 |
ENST00000560752.1
|
ADNP2
|
ADNP homeobox 2 |
| chr2_-_175462934 | 0.04 |
ENST00000392546.2
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr18_-_72265035 | 0.04 |
ENST00000585279.1
ENST00000580048.1 |
LINC00909
|
long intergenic non-protein coding RNA 909 |
| chr1_+_173684047 | 0.04 |
ENST00000546011.1
ENST00000209884.4 |
KLHL20
|
kelch-like family member 20 |
| chr5_+_118406796 | 0.04 |
ENST00000503802.1
|
DMXL1
|
Dmx-like 1 |
| chr16_-_67260691 | 0.04 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr3_-_45883558 | 0.04 |
ENST00000445698.1
ENST00000296135.6 |
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr1_+_154193643 | 0.04 |
ENST00000456325.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr8_-_38034234 | 0.04 |
ENST00000311351.4
|
LSM1
|
LSM1 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr2_+_38152462 | 0.04 |
ENST00000354545.2
|
RMDN2
|
regulator of microtubule dynamics 2 |
| chr1_+_16083123 | 0.04 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr21_-_27107344 | 0.04 |
ENST00000457143.2
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr6_+_30524663 | 0.04 |
ENST00000376560.3
|
PRR3
|
proline rich 3 |
| chr5_+_80597453 | 0.04 |
ENST00000438268.2
|
ZCCHC9
|
zinc finger, CCHC domain containing 9 |
| chr4_+_76439095 | 0.04 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr12_+_9102632 | 0.04 |
ENST00000539240.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr4_+_175205100 | 0.04 |
ENST00000515299.1
|
CEP44
|
centrosomal protein 44kDa |
| chrX_+_70435044 | 0.04 |
ENST00000374029.1
ENST00000374022.3 ENST00000447581.1 |
GJB1
|
gap junction protein, beta 1, 32kDa |
| chr15_+_77713299 | 0.04 |
ENST00000559099.1
|
HMG20A
|
high mobility group 20A |
| chr6_-_31619892 | 0.04 |
ENST00000454165.1
ENST00000428326.1 ENST00000452994.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr5_+_134094461 | 0.04 |
ENST00000452510.2
ENST00000354283.4 |
DDX46
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr11_+_113185571 | 0.04 |
ENST00000455306.1
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr8_+_29953163 | 0.04 |
ENST00000518192.1
|
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr4_-_156298028 | 0.04 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr12_-_66524482 | 0.04 |
ENST00000446587.2
ENST00000266604.2 |
LLPH
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
Network of associatons between targets according to the STRING database.
First level regulatory network of ETS1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.0 | 0.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.2 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.0 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.1 | GO:0044053 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.0 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.0 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.0 | 0.2 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.0 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.1 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.0 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.0 | GO:0035350 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.0 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.0 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.0 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |