Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
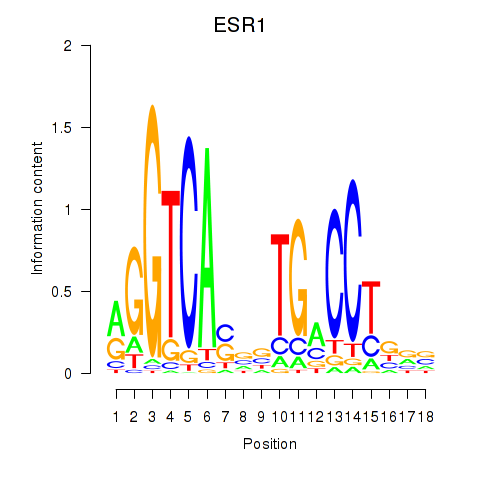
Results for ESR1
Z-value: 0.13
Transcription factors associated with ESR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESR1
|
ENSG00000091831.17 | estrogen receptor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ESR1 | hg19_v2_chr6_+_152130240_152130274 | 0.96 | 4.2e-02 | Click! |
Activity profile of ESR1 motif
Sorted Z-values of ESR1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_144099795 | 0.07 |
ENST00000522060.1
ENST00000517833.1 ENST00000502167.2 ENST00000518831.1 |
RP11-273G15.2
|
RP11-273G15.2 |
| chr22_+_23029188 | 0.06 |
ENST00000390305.2
|
IGLV3-25
|
immunoglobulin lambda variable 3-25 |
| chr8_+_120885949 | 0.06 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr10_+_115438920 | 0.05 |
ENST00000429617.1
ENST00000369331.4 |
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr2_+_242681835 | 0.04 |
ENST00000437164.1
ENST00000454048.1 ENST00000417686.1 |
D2HGDH
|
D-2-hydroxyglutarate dehydrogenase |
| chr21_-_47352477 | 0.04 |
ENST00000593412.1
|
PRED62
|
Uncharacterized protein |
| chr1_-_16556038 | 0.04 |
ENST00000375605.2
|
C1orf134
|
chromosome 1 open reading frame 134 |
| chr3_+_195413160 | 0.04 |
ENST00000599448.1
|
LINC00969
|
long intergenic non-protein coding RNA 969 |
| chr12_+_123874589 | 0.04 |
ENST00000437502.1
|
SETD8
|
SET domain containing (lysine methyltransferase) 8 |
| chr14_+_94577074 | 0.04 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr19_-_5624057 | 0.04 |
ENST00000590262.1
|
SAFB2
|
scaffold attachment factor B2 |
| chr19_+_36603662 | 0.03 |
ENST00000586670.1
|
OVOL3
|
ovo-like zinc finger 3 |
| chr12_-_30907862 | 0.03 |
ENST00000541765.1
ENST00000537108.1 |
CAPRIN2
|
caprin family member 2 |
| chr11_-_12030681 | 0.03 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr13_-_36705425 | 0.03 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr3_-_117716418 | 0.03 |
ENST00000484092.1
|
RP11-384F7.2
|
RP11-384F7.2 |
| chr12_-_30907822 | 0.03 |
ENST00000540436.1
|
CAPRIN2
|
caprin family member 2 |
| chr3_+_129207033 | 0.03 |
ENST00000507221.1
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr19_-_59010565 | 0.03 |
ENST00000594786.1
|
SLC27A5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr9_+_130478345 | 0.03 |
ENST00000373289.3
ENST00000393748.4 |
TTC16
|
tetratricopeptide repeat domain 16 |
| chr1_-_111970353 | 0.03 |
ENST00000369732.3
|
OVGP1
|
oviductal glycoprotein 1, 120kDa |
| chr17_+_63096903 | 0.03 |
ENST00000582940.1
|
RP11-160O5.1
|
RP11-160O5.1 |
| chr12_-_30907749 | 0.03 |
ENST00000542550.1
ENST00000540584.1 |
CAPRIN2
|
caprin family member 2 |
| chr13_-_24471194 | 0.03 |
ENST00000382137.3
ENST00000382057.3 |
C1QTNF9B
|
C1q and tumor necrosis factor related protein 9B |
| chr19_+_1261106 | 0.03 |
ENST00000588411.1
|
CIRBP
|
cold inducible RNA binding protein |
| chr4_+_76871883 | 0.03 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr13_+_103451548 | 0.03 |
ENST00000419638.1
|
BIVM
|
basic, immunoglobulin-like variable motif containing |
| chr17_+_73629500 | 0.03 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr22_-_32034376 | 0.03 |
ENST00000431201.1
|
PISD
|
phosphatidylserine decarboxylase |
| chr7_+_106415457 | 0.03 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr3_-_196014520 | 0.03 |
ENST00000441879.1
ENST00000292823.2 ENST00000411591.1 ENST00000431016.1 ENST00000443555.1 |
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr19_+_1452188 | 0.03 |
ENST00000587149.1
|
APC2
|
adenomatosis polyposis coli 2 |
| chr9_-_127905736 | 0.03 |
ENST00000336505.6
ENST00000373549.4 |
SCAI
|
suppressor of cancer cell invasion |
| chr2_-_98612379 | 0.03 |
ENST00000425805.2
|
TMEM131
|
transmembrane protein 131 |
| chr15_-_83621435 | 0.02 |
ENST00000450735.2
ENST00000426485.1 ENST00000399166.2 ENST00000304231.8 |
HOMER2
|
homer homolog 2 (Drosophila) |
| chr7_+_30068260 | 0.02 |
ENST00000440706.2
|
PLEKHA8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr9_+_33240157 | 0.02 |
ENST00000379721.3
|
SPINK4
|
serine peptidase inhibitor, Kazal type 4 |
| chr1_+_55446465 | 0.02 |
ENST00000371268.3
|
TMEM61
|
transmembrane protein 61 |
| chr1_+_1266654 | 0.02 |
ENST00000339381.5
|
TAS1R3
|
taste receptor, type 1, member 3 |
| chr6_+_28092338 | 0.02 |
ENST00000340487.4
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr1_-_85156417 | 0.02 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr15_-_72462384 | 0.02 |
ENST00000568594.1
|
GRAMD2
|
GRAM domain containing 2 |
| chr8_+_96281045 | 0.02 |
ENST00000521905.1
|
KB-1047C11.2
|
KB-1047C11.2 |
| chr14_-_91884115 | 0.02 |
ENST00000389857.6
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr14_-_75536182 | 0.02 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr16_-_3137080 | 0.02 |
ENST00000574387.1
ENST00000571404.1 |
RP11-473M20.9
|
RP11-473M20.9 |
| chr14_-_91884150 | 0.02 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr17_+_65374075 | 0.02 |
ENST00000581322.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr12_-_56359802 | 0.02 |
ENST00000548803.1
ENST00000449260.2 ENST00000550447.1 ENST00000547137.1 ENST00000546543.1 ENST00000550464.1 |
PMEL
|
premelanosome protein |
| chr2_-_241396131 | 0.02 |
ENST00000404327.3
|
AC110619.2
|
Uncharacterized protein |
| chr15_+_90931450 | 0.02 |
ENST00000268182.5
ENST00000560738.1 ENST00000560418.1 |
IQGAP1
|
IQ motif containing GTPase activating protein 1 |
| chr11_+_8704298 | 0.02 |
ENST00000531978.1
ENST00000524496.1 ENST00000532359.1 ENST00000530022.1 |
RPL27A
|
ribosomal protein L27a |
| chr12_-_88535747 | 0.02 |
ENST00000309041.7
|
CEP290
|
centrosomal protein 290kDa |
| chr10_+_14880364 | 0.02 |
ENST00000441647.1
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr19_+_1942453 | 0.02 |
ENST00000591752.1
|
CSNK1G2
|
casein kinase 1, gamma 2 |
| chr4_-_668108 | 0.02 |
ENST00000304312.4
|
ATP5I
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E |
| chr19_-_10445399 | 0.02 |
ENST00000592945.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr11_+_8704748 | 0.02 |
ENST00000526562.1
ENST00000525981.1 |
RPL27A
|
ribosomal protein L27a |
| chr18_+_21529811 | 0.02 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr3_+_63897605 | 0.02 |
ENST00000487717.1
|
ATXN7
|
ataxin 7 |
| chr8_-_143867946 | 0.02 |
ENST00000301263.4
|
LY6D
|
lymphocyte antigen 6 complex, locus D |
| chr19_+_42381173 | 0.02 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr3_-_138312971 | 0.02 |
ENST00000485115.1
ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70
|
centrosomal protein 70kDa |
| chr11_+_1891380 | 0.02 |
ENST00000429923.1
ENST00000418975.1 ENST00000406638.2 |
LSP1
|
lymphocyte-specific protein 1 |
| chr22_+_22676808 | 0.02 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr19_+_23257988 | 0.02 |
ENST00000594318.1
ENST00000600223.1 ENST00000593635.1 |
CTD-2291D10.4
ZNF730
|
CTD-2291D10.4 zinc finger protein 730 |
| chr6_-_159420780 | 0.02 |
ENST00000449822.1
|
RSPH3
|
radial spoke 3 homolog (Chlamydomonas) |
| chr11_-_12030746 | 0.02 |
ENST00000533813.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr2_-_231090344 | 0.02 |
ENST00000540870.1
ENST00000416610.1 |
SP110
|
SP110 nuclear body protein |
| chr10_-_27389320 | 0.02 |
ENST00000436985.2
|
ANKRD26
|
ankyrin repeat domain 26 |
| chr3_+_169684553 | 0.02 |
ENST00000337002.4
ENST00000480708.1 |
SEC62
|
SEC62 homolog (S. cerevisiae) |
| chr1_-_223853425 | 0.02 |
ENST00000366873.2
ENST00000419193.2 |
CAPN8
|
calpain 8 |
| chr14_+_52164675 | 0.02 |
ENST00000555936.1
|
FRMD6
|
FERM domain containing 6 |
| chr2_+_204103733 | 0.02 |
ENST00000443941.1
|
CYP20A1
|
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr16_-_19725899 | 0.02 |
ENST00000567367.1
|
KNOP1
|
lysine-rich nucleolar protein 1 |
| chr19_-_43702231 | 0.02 |
ENST00000597374.1
ENST00000599371.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr2_+_233243233 | 0.02 |
ENST00000392027.2
|
ALPP
|
alkaline phosphatase, placental |
| chr16_-_74734672 | 0.02 |
ENST00000306247.7
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr16_+_66914264 | 0.02 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr15_-_52404921 | 0.02 |
ENST00000561198.1
ENST00000260442.3 |
BCL2L10
|
BCL2-like 10 (apoptosis facilitator) |
| chr12_+_97300995 | 0.02 |
ENST00000266742.4
ENST00000429527.2 ENST00000554226.1 ENST00000557478.1 ENST00000557092.1 ENST00000411739.2 ENST00000553609.1 |
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr3_-_52002194 | 0.02 |
ENST00000466412.1
|
PCBP4
|
poly(rC) binding protein 4 |
| chr21_-_40817645 | 0.02 |
ENST00000438404.1
ENST00000358268.2 ENST00000411566.1 ENST00000451131.1 ENST00000418018.1 ENST00000415863.1 ENST00000426783.1 ENST00000288350.3 ENST00000485895.2 ENST00000448288.2 ENST00000456017.1 ENST00000434281.1 |
LCA5L
|
Leber congenital amaurosis 5-like |
| chr8_-_131028660 | 0.02 |
ENST00000401979.2
ENST00000517654.1 ENST00000522361.1 ENST00000518167.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr19_-_2783255 | 0.02 |
ENST00000589251.1
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr19_+_19516561 | 0.02 |
ENST00000457895.2
|
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr4_+_77941685 | 0.02 |
ENST00000506731.1
|
SEPT11
|
septin 11 |
| chr1_-_223853348 | 0.02 |
ENST00000366872.5
|
CAPN8
|
calpain 8 |
| chr12_+_16500571 | 0.02 |
ENST00000543076.1
ENST00000396210.3 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr14_+_52456193 | 0.02 |
ENST00000261700.3
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr3_+_9774164 | 0.02 |
ENST00000426583.1
|
BRPF1
|
bromodomain and PHD finger containing, 1 |
| chr3_-_138313161 | 0.02 |
ENST00000489254.1
ENST00000474781.1 ENST00000462419.1 ENST00000264982.3 |
CEP70
|
centrosomal protein 70kDa |
| chr5_+_179105615 | 0.02 |
ENST00000514383.1
|
CANX
|
calnexin |
| chr2_-_69664549 | 0.02 |
ENST00000450796.2
ENST00000484177.1 |
NFU1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr1_-_38218577 | 0.02 |
ENST00000540011.1
|
EPHA10
|
EPH receptor A10 |
| chr12_+_100967420 | 0.02 |
ENST00000266754.5
ENST00000547754.1 |
GAS2L3
|
growth arrest-specific 2 like 3 |
| chr11_+_111750206 | 0.02 |
ENST00000530214.1
ENST00000530799.1 |
C11orf1
|
chromosome 11 open reading frame 1 |
| chr6_+_144164455 | 0.02 |
ENST00000367576.5
|
LTV1
|
LTV1 homolog (S. cerevisiae) |
| chr11_+_75526212 | 0.02 |
ENST00000356136.3
|
UVRAG
|
UV radiation resistance associated |
| chr7_-_92146729 | 0.02 |
ENST00000541751.1
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr9_+_129987488 | 0.02 |
ENST00000446764.1
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr19_+_52800410 | 0.02 |
ENST00000595962.1
ENST00000598016.1 ENST00000334564.7 ENST00000490272.1 |
ZNF480
|
zinc finger protein 480 |
| chr1_+_209848749 | 0.02 |
ENST00000367029.4
|
G0S2
|
G0/G1switch 2 |
| chr19_+_58790314 | 0.02 |
ENST00000196548.5
ENST00000608843.1 |
ZNF8
ZNF8
|
Zinc finger protein 8 zinc finger protein 8 |
| chr1_+_245133656 | 0.02 |
ENST00000366521.3
|
EFCAB2
|
EF-hand calcium binding domain 2 |
| chr11_+_64008443 | 0.02 |
ENST00000309366.4
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr3_-_122512619 | 0.02 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr17_-_79359154 | 0.02 |
ENST00000572077.1
|
RP11-1055B8.3
|
RP11-1055B8.3 |
| chr15_-_72462661 | 0.02 |
ENST00000570275.1
|
GRAMD2
|
GRAM domain containing 2 |
| chr9_+_136243117 | 0.02 |
ENST00000426926.2
ENST00000371957.3 |
C9orf96
|
chromosome 9 open reading frame 96 |
| chr16_+_70207686 | 0.02 |
ENST00000541793.2
ENST00000314151.8 ENST00000565806.1 ENST00000569347.2 ENST00000536907.2 |
CLEC18C
|
C-type lectin domain family 18, member C |
| chr12_-_131478468 | 0.02 |
ENST00000536673.1
ENST00000542980.1 |
RP11-76C10.5
|
RP11-76C10.5 |
| chr15_+_81225699 | 0.02 |
ENST00000560027.1
|
KIAA1199
|
KIAA1199 |
| chr19_-_37157713 | 0.02 |
ENST00000592829.1
ENST00000591370.1 ENST00000588268.1 ENST00000360357.4 |
ZNF461
|
zinc finger protein 461 |
| chr9_+_110046334 | 0.02 |
ENST00000416373.2
|
RAD23B
|
RAD23 homolog B (S. cerevisiae) |
| chr3_-_15469045 | 0.02 |
ENST00000450816.2
|
METTL6
|
methyltransferase like 6 |
| chr16_-_1275257 | 0.02 |
ENST00000234798.4
|
TPSG1
|
tryptase gamma 1 |
| chr4_+_128982430 | 0.02 |
ENST00000512292.1
ENST00000508819.1 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr19_+_10222189 | 0.02 |
ENST00000321826.4
|
P2RY11
|
purinergic receptor P2Y, G-protein coupled, 11 |
| chr14_+_52456327 | 0.02 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr17_+_41005283 | 0.02 |
ENST00000592999.1
|
AOC3
|
amine oxidase, copper containing 3 |
| chr11_-_69294647 | 0.02 |
ENST00000542064.1
|
AP000439.3
|
AP000439.3 |
| chr18_+_5238549 | 0.02 |
ENST00000580684.1
|
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr16_+_1306093 | 0.02 |
ENST00000211076.3
|
TPSD1
|
tryptase delta 1 |
| chr19_+_42824511 | 0.02 |
ENST00000601644.1
|
TMEM145
|
transmembrane protein 145 |
| chr13_-_52027134 | 0.02 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr7_+_101460882 | 0.02 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr14_+_103566665 | 0.02 |
ENST00000559116.1
|
EXOC3L4
|
exocyst complex component 3-like 4 |
| chr22_-_32034409 | 0.02 |
ENST00000429683.1
|
PISD
|
phosphatidylserine decarboxylase |
| chr6_-_2903514 | 0.02 |
ENST00000380698.4
|
SERPINB9
|
serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
| chr11_+_64008525 | 0.02 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr19_+_38880695 | 0.02 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr4_-_186347099 | 0.02 |
ENST00000505357.1
ENST00000264689.6 |
UFSP2
|
UFM1-specific peptidase 2 |
| chr3_-_47205457 | 0.01 |
ENST00000409792.3
|
SETD2
|
SET domain containing 2 |
| chr5_+_667759 | 0.01 |
ENST00000594226.1
|
AC026740.1
|
Uncharacterized protein |
| chr2_+_11679963 | 0.01 |
ENST00000263834.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr8_-_131028641 | 0.01 |
ENST00000523509.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr19_+_20011775 | 0.01 |
ENST00000592245.1
ENST00000592160.1 ENST00000343769.5 |
AC007204.2
ZNF93
|
AC007204.2 zinc finger protein 93 |
| chr12_+_16500599 | 0.01 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr8_-_123793048 | 0.01 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr15_+_90792760 | 0.01 |
ENST00000339615.5
ENST00000438251.1 |
TTLL13
|
tubulin tyrosine ligase-like family, member 13 |
| chr6_-_41006928 | 0.01 |
ENST00000244565.3
|
UNC5CL
|
unc-5 homolog C (C. elegans)-like |
| chr8_+_30013813 | 0.01 |
ENST00000221114.3
|
DCTN6
|
dynactin 6 |
| chr18_+_47901408 | 0.01 |
ENST00000398452.2
ENST00000417656.2 ENST00000488454.1 ENST00000494518.1 |
SKA1
|
spindle and kinetochore associated complex subunit 1 |
| chr22_-_23922410 | 0.01 |
ENST00000249053.3
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr2_+_204103663 | 0.01 |
ENST00000356079.4
ENST00000429815.2 |
CYP20A1
|
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr10_-_27389392 | 0.01 |
ENST00000376087.4
|
ANKRD26
|
ankyrin repeat domain 26 |
| chr4_-_682769 | 0.01 |
ENST00000507165.1
|
MFSD7
|
major facilitator superfamily domain containing 7 |
| chr7_+_26241325 | 0.01 |
ENST00000456948.1
ENST00000409747.1 |
CBX3
|
chromobox homolog 3 |
| chr18_+_56531584 | 0.01 |
ENST00000590287.1
|
ZNF532
|
zinc finger protein 532 |
| chr3_-_148804275 | 0.01 |
ENST00000392912.2
ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF
|
helicase-like transcription factor |
| chr19_+_39936207 | 0.01 |
ENST00000594729.1
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr1_+_154193325 | 0.01 |
ENST00000428931.1
ENST00000441890.1 ENST00000271877.7 ENST00000412596.1 ENST00000368504.1 ENST00000437652.1 |
UBAP2L
|
ubiquitin associated protein 2-like |
| chr7_+_92861653 | 0.01 |
ENST00000251739.5
ENST00000305866.5 ENST00000544910.1 ENST00000541136.1 ENST00000458530.1 ENST00000535481.1 ENST00000317751.6 |
CCDC132
|
coiled-coil domain containing 132 |
| chr7_+_6071007 | 0.01 |
ENST00000409061.1
|
ANKRD61
|
ankyrin repeat domain 61 |
| chr16_+_71879861 | 0.01 |
ENST00000427980.2
ENST00000568581.1 |
ATXN1L
IST1
|
ataxin 1-like increased sodium tolerance 1 homolog (yeast) |
| chr15_-_49102781 | 0.01 |
ENST00000558591.1
|
CEP152
|
centrosomal protein 152kDa |
| chr17_-_39677971 | 0.01 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr3_+_38323785 | 0.01 |
ENST00000466887.1
ENST00000448498.1 |
SLC22A14
|
solute carrier family 22, member 14 |
| chr7_+_142982023 | 0.01 |
ENST00000359333.3
ENST00000409244.1 ENST00000409541.1 ENST00000410004.1 |
TMEM139
|
transmembrane protein 139 |
| chr19_-_2090131 | 0.01 |
ENST00000591326.1
|
MOB3A
|
MOB kinase activator 3A |
| chr16_+_28857957 | 0.01 |
ENST00000567536.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr10_-_48332197 | 0.01 |
ENST00000454672.1
|
RP11-463P17.1
|
RP11-463P17.1 |
| chr16_+_83841448 | 0.01 |
ENST00000433866.2
|
HSBP1
|
heat shock factor binding protein 1 |
| chr9_+_140125385 | 0.01 |
ENST00000361134.2
|
SLC34A3
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 3 |
| chr10_+_76586348 | 0.01 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr1_+_26348259 | 0.01 |
ENST00000374280.3
|
EXTL1
|
exostosin-like glycosyltransferase 1 |
| chr8_-_41522779 | 0.01 |
ENST00000522231.1
ENST00000314214.8 ENST00000348036.4 ENST00000457297.1 ENST00000522543.1 |
ANK1
|
ankyrin 1, erythrocytic |
| chr19_+_46002868 | 0.01 |
ENST00000396735.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr18_+_9913977 | 0.01 |
ENST00000400000.2
ENST00000340541.4 |
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
| chr2_-_42588289 | 0.01 |
ENST00000468711.1
ENST00000463055.1 |
COX7A2L
|
cytochrome c oxidase subunit VIIa polypeptide 2 like |
| chr17_-_76274572 | 0.01 |
ENST00000374945.1
|
RP11-219G17.4
|
RP11-219G17.4 |
| chr1_+_179851999 | 0.01 |
ENST00000527391.1
|
TOR1AIP1
|
torsin A interacting protein 1 |
| chr2_+_27282134 | 0.01 |
ENST00000441931.1
|
AGBL5
|
ATP/GTP binding protein-like 5 |
| chr12_+_122356488 | 0.01 |
ENST00000397454.2
|
WDR66
|
WD repeat domain 66 |
| chr22_-_23922448 | 0.01 |
ENST00000438703.1
ENST00000330377.2 |
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr8_-_53626974 | 0.01 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chr12_-_42877726 | 0.01 |
ENST00000548696.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr5_-_175395283 | 0.01 |
ENST00000513482.1
ENST00000265097.4 |
THOC3
|
THO complex 3 |
| chr22_+_39417118 | 0.01 |
ENST00000216099.8
|
APOBEC3D
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D |
| chr2_+_219283815 | 0.01 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr17_+_38673270 | 0.01 |
ENST00000578280.1
|
RP5-1028K7.2
|
RP5-1028K7.2 |
| chr19_-_36001386 | 0.01 |
ENST00000461300.1
|
DMKN
|
dermokine |
| chr5_+_131892815 | 0.01 |
ENST00000453394.1
|
RAD50
|
RAD50 homolog (S. cerevisiae) |
| chr7_-_30544405 | 0.01 |
ENST00000409390.1
ENST00000409144.1 ENST00000005374.6 ENST00000409436.1 ENST00000275428.4 |
GGCT
|
gamma-glutamylcyclotransferase |
| chr11_+_69924679 | 0.01 |
ENST00000531604.1
|
ANO1
|
anoctamin 1, calcium activated chloride channel |
| chr17_+_75446629 | 0.01 |
ENST00000588958.2
ENST00000586128.1 |
SEPT9
|
septin 9 |
| chr2_+_131862872 | 0.01 |
ENST00000439822.2
|
PLEKHB2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr19_-_13068012 | 0.01 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr17_+_46985823 | 0.01 |
ENST00000508468.2
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z |
| chr6_-_24358264 | 0.01 |
ENST00000378454.3
|
DCDC2
|
doublecortin domain containing 2 |
| chr17_-_41277467 | 0.01 |
ENST00000494123.1
ENST00000346315.3 ENST00000309486.4 ENST00000468300.1 ENST00000354071.3 ENST00000352993.3 ENST00000471181.2 |
BRCA1
|
breast cancer 1, early onset |
| chr11_+_119019722 | 0.01 |
ENST00000307417.3
|
ABCG4
|
ATP-binding cassette, sub-family G (WHITE), member 4 |
| chr15_+_89182178 | 0.01 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr18_-_3247084 | 0.01 |
ENST00000609924.1
|
RP13-270P17.3
|
RP13-270P17.3 |
| chr1_-_13840483 | 0.01 |
ENST00000376085.3
|
LRRC38
|
leucine rich repeat containing 38 |
| chr8_+_17780483 | 0.01 |
ENST00000517730.1
ENST00000518537.1 ENST00000523055.1 ENST00000519253.1 |
PCM1
|
pericentriolar material 1 |
| chr4_+_128982490 | 0.01 |
ENST00000394288.3
ENST00000432347.2 ENST00000264584.5 ENST00000441387.1 ENST00000427266.1 ENST00000354456.3 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr5_-_40798473 | 0.01 |
ENST00000354209.3
|
PRKAA1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit |
| chr7_+_149535455 | 0.01 |
ENST00000223210.4
ENST00000460379.1 |
ZNF862
|
zinc finger protein 862 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ESR1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.0 | 0.0 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0016898 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |